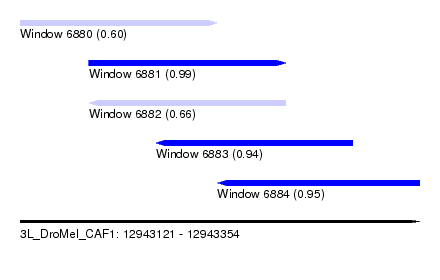
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,943,121 – 12,943,354 |
| Length | 233 |
| Max. P | 0.991055 |

| Location | 12,943,121 – 12,943,236 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -20.61 |
| Consensus MFE | -16.43 |
| Energy contribution | -17.75 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12943121 115 + 23771897 ACAACACAUGCUACUAAUUUGCGAAAUUAACGUGUUUGUUUUGGCCAUUCUCUAAAUAAAUUACCCCCAAA-AAACAAAAUACAG-G---AAGAAAACGAGCAGCCAAAGGUAAAGGCAA ........((((..(((((.....)))))..((((((.((((((......................)))))-)....))))))..-.---.........))))(((.........))).. ( -14.55) >DroSec_CAF1 60378 115 + 1 ACAACACAUGCUACUAAUUUGCGAAAUUAACGUGUUUGUUUUGGCCGCUCUCUAAAUAAAUUACCUCCAAAAAAACAAAAUACAG-G---AA-AAAACGAGCGGCCAAAGGUAAAGGCAA ........((((.....(((((.((((......)))).((((((((((((..(((.....)))..(((................)-)---).-.....))))))))))))))))))))). ( -24.59) >DroSim_CAF1 64725 115 + 1 ACAACACAUGCUACUAAUUUGCGAAAUUAACGUGUUUGUUUUGGCCGCUCUCUAAAUAAAUUACCUCCAAAAAAACAAAAUACAG-G---AA-AAAACGAGCGGCCAAAGGUAAAGGCAA ........((((.....(((((.((((......)))).((((((((((((..(((.....)))..(((................)-)---).-.....))))))))))))))))))))). ( -24.59) >DroEre_CAF1 66624 110 + 1 ACAACACAUGCUACUAAUUUGCGAAAUUAACGUGUUUGUUUUGGCCGCUGUGUAAAUAAAUUACACCCAAAAA------ACACAGAG---AA-AAAACGCCCGGCCAAAGGUAAAGGCAA ........((((.....(((((.((((......)))).(((((((((((((((....................------)))))).(---..-....)...)))))))))))))))))). ( -21.15) >DroYak_CAF1 64193 113 + 1 ACAACACAUGCUACUAAUUUGCGAAAUUAACGUGUUUGUUUUGGCCGCUCUCUAAAUAAAUUACCCCCAAAAA------AUACAGAGAAGAA-AAAACGCCCGGCCAAAGGUAAAGGCAA ........((((.....(((((.((((......)))).(((((((((.(((((....................------....))))).(..-....)...)))))))))))))))))). ( -18.16) >consensus ACAACACAUGCUACUAAUUUGCGAAAUUAACGUGUUUGUUUUGGCCGCUCUCUAAAUAAAUUACCCCCAAAAAAACAAAAUACAG_G___AA_AAAACGAGCGGCCAAAGGUAAAGGCAA ........((((.....(((((.((((......)))).((((((((((((................................................))))))))))))))))))))). (-16.43 = -17.75 + 1.32)



| Location | 12,943,161 – 12,943,276 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -21.07 |
| Energy contribution | -22.59 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12943161 115 + 23771897 UUGGCCAUUCUCUAAAUAAAUUACCCCCAAA-AAACAAAAUACAG-G---AAGAAAACGAGCAGCCAAAGGUAAAGGCAAUGUCCAUAAAUUCGCAUCGAAAGCGAAUUCCGAGCUGCAA ...............................-.............-.---..........(((((....((....((......))...(((((((.......)))))))))..))))).. ( -17.30) >DroSec_CAF1 60418 115 + 1 UUGGCCGCUCUCUAAAUAAAUUACCUCCAAAAAAACAAAAUACAG-G---AA-AAAACGAGCGGCCAAAGGUAAAGGCAAUGGCCAUAAAUUCGCAUCGAAAGCGAAUUCCGAGCUGCAA ((((((((((..(((.....)))..(((................)-)---).-.....)))))))))).((....(((....)))...(((((((.......)))))))))......... ( -30.39) >DroSim_CAF1 64765 115 + 1 UUGGCCGCUCUCUAAAUAAAUUACCUCCAAAAAAACAAAAUACAG-G---AA-AAAACGAGCGGCCAAAGGUAAAGGCAAUGGCCAUAAAUUCGCAUCGAAAGCGAAUUCCGAGCUGCAA ((((((((((..(((.....)))..(((................)-)---).-.....)))))))))).((....(((....)))...(((((((.......)))))))))......... ( -30.39) >DroEre_CAF1 66664 110 + 1 UUGGCCGCUGUGUAAAUAAAUUACACCCAAAAA------ACACAGAG---AA-AAAACGCCCGGCCAAAGGUAAAGGCAAUGGCCAUAAAUUCGCAUCGAAAGCGAAUUCCGAGCUGCAA .((((((((((((....................------))))))..---..-.....(((..(((...)))...)))..))))))..(((((((.......)))))))........... ( -28.25) >DroYak_CAF1 64233 113 + 1 UUGGCCGCUCUCUAAAUAAAUUACCCCCAAAAA------AUACAGAGAAGAA-AAAACGCCCGGCCAAAGGUAAAGGCAAUGGCCAUAAAUUCGCAUCGAAAGCGAAUUCCGAGCUGCAA ...((.((((.......................------.............-.........(((((...((....))..)))))...(((((((.......)))))))..)))).)).. ( -25.80) >consensus UUGGCCGCUCUCUAAAUAAAUUACCCCCAAAAAAACAAAAUACAG_G___AA_AAAACGAGCGGCCAAAGGUAAAGGCAAUGGCCAUAAAUUCGCAUCGAAAGCGAAUUCCGAGCUGCAA ((((((((((................................................)))))))))).((....(((....)))...(((((((.......)))))))))......... (-21.07 = -22.59 + 1.52)

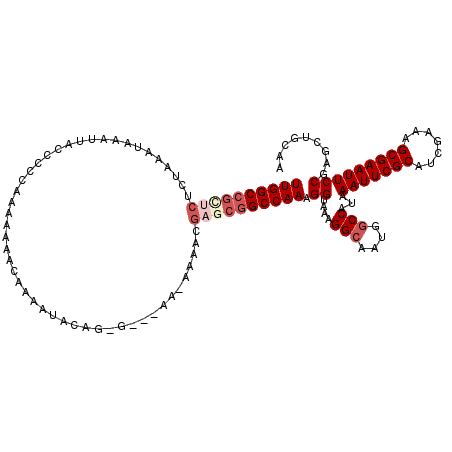

| Location | 12,943,161 – 12,943,276 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12943161 115 - 23771897 UUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGACAUUGCCUUUACCUUUGGCUGCUCGUUUUCUU---C-CUGUAUUUUGUUU-UUUGGGGGUAAUUUAUUUAGAGAAUGGCCAA ..((((((..((((((((.......))))))))..((......)).........))))))(((((((((.---.-..(((..(((..(-.....)..)))..)))..))))))))).... ( -27.80) >DroSec_CAF1 60418 115 - 1 UUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGCUCGUUUU-UU---C-CUGUAUUUUGUUUUUUUGGAGGUAAUUUAUUUAGAGAGCGGCCAA ..((((((..((((((((.......))))))))..)))...)))........((((((((((....(-((---(-(................)))))(((.....)))..)))))))))) ( -34.79) >DroSim_CAF1 64765 115 - 1 UUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGCUCGUUUU-UU---C-CUGUAUUUUGUUUUUUUGGAGGUAAUUUAUUUAGAGAGCGGCCAA ..((((((..((((((((.......))))))))..)))...)))........((((((((((....(-((---(-(................)))))(((.....)))..)))))))))) ( -34.79) >DroEre_CAF1 66664 110 - 1 UUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGGGCGUUUU-UU---CUCUGUGU------UUUUUGGGUGUAAUUUAUUUACACAGCGGCCAA .(((.((((.((((((((.......))))))))..(((((..(.......)..))))))))).....-..---........------.......((((((.....)))))).)))..... ( -32.40) >DroYak_CAF1 64233 113 - 1 UUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGGGCGUUUU-UUCUUCUCUGUAU------UUUUUGGGGGUAAUUUAUUUAGAGAGCGGCCAA .(((.((((.((((((((.......))))))))..(((((..(.......)..))))))))).....-....((((((.((------...(((....)))...)).)))))))))..... ( -31.00) >consensus UUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGCUCGUUUU_UU___C_CUGUAUUUUGUUUUUUUGGGGGUAAUUUAUUUAGAGAGCGGCCAA ..((.((((.((((((((.......))))))))..(((....))).(((((((..(..................................)..)))))))..........)))).))... (-22.48 = -22.88 + 0.40)

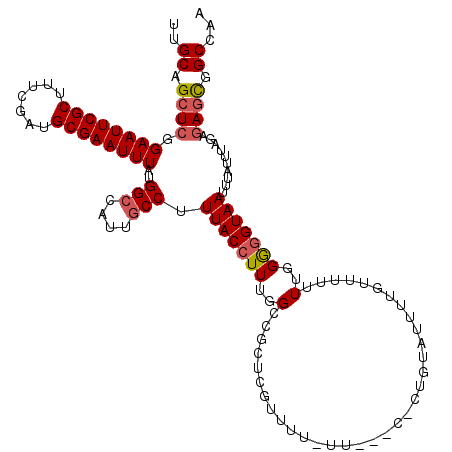
| Location | 12,943,200 – 12,943,315 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -32.76 |
| Consensus MFE | -26.76 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12943200 115 - 23771897 UCCUU-UUUGUGGGCAGUAAACAAGGCAACCAACCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGACAUUGCCUUUACCUUUGGCUGCUCGUUUUCUU---C-CUGUA .....-.....(((((((....(((((((.((((.....))))..(..((((((((((.......)))))))).))..).)))))))........)))))))........---.-..... ( -30.10) >DroSec_CAF1 60458 111 - 1 UCCUUUUUUGUGGGCAGUAAACAAGGCAA----CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGCUCGUUUU-UU---C-CUGUA .((........))((((.((((...((((----(.....)))))(((...((((((((.......))))))))..(((((..(.......)..)))))))).)))).-..---.-)))). ( -32.10) >DroSim_CAF1 64805 110 - 1 UCCUU-UUUGUGGGCAGUAAACAAGGCAA----CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGCUCGUUUU-UU---C-CUGUA .((..-.....))((((.((((...((((----(.....)))))(((...((((((((.......))))))))..(((((..(.......)..)))))))).)))).-..---.-)))). ( -31.80) >DroEre_CAF1 66698 112 - 1 UUCUUUUUUGUGGGCAGUAAACAAGGCAA----CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGGGCGUUUU-UU---CUCUGUG .............((((.((((...((((----(.....))))).((((.((((((((.......))))))))..(((((..(.......)..))))))))))))).-..---..)))). ( -35.00) >DroYak_CAF1 64267 115 - 1 UUCUUUUUUGUGGGCAGUAAACAAGGCAA----CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGGGCGUUUU-UUCUUCUCUGUA ...........(((.((.((((...((((----(.....))))).((((.((((((((.......))))))))..(((((..(.......)..))))))))))))).-..)).))).... ( -34.80) >consensus UCCUUUUUUGUGGGCAGUAAACAAGGCAA____CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCAUUGCCUUUACCUUUGGCCGCUCGUUUU_UU___C_CUGUA .........((((.(((.....(((((((.....(((.(((....)))..((((((((.......)))))))).)))...))))))).....))).)))).................... (-26.76 = -27.00 + 0.24)

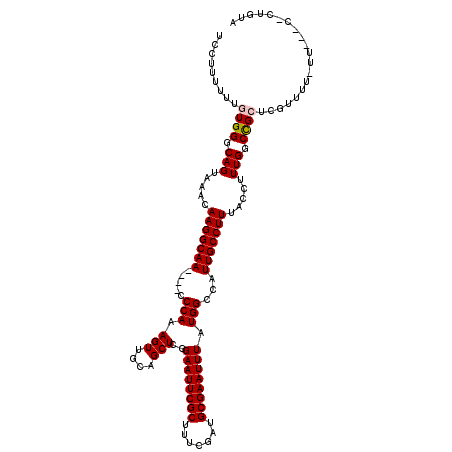
| Location | 12,943,236 – 12,943,354 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -30.36 |
| Energy contribution | -30.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12943236 118 - 23771897 AGUUGGCUGUAAUUU-UUGGUCAUGAUAUUGAGAAGGGGAUCCUU-UUUGUGGGCAGUAAACAAGGCAACCAACCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGACA .((((((((((((((-(.(((.........(((((((....))))-))).(((...((.......))..)))))).))))))))))))..((((((((.......))))))))...))). ( -35.30) >DroSec_CAF1 60493 116 - 1 AGUUGGCUGUAAUUUUUUGGCCAUGGUAUUGAGAAGGGGAUCCUUUUUUGUGGGCAGUAAACAAGGCAA----CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCA .................(((((((...((.(((((((....))))))).))((((.(....)...((((----(.....))))).)))).((((((((.......))))))))))))))) ( -34.60) >DroSim_CAF1 64840 114 - 1 AGUUGGCUGUAAUUU-UUGGCCAUGGUAUUGAGAAGGGGAUCCUU-UUUGUGGGCAGUAAACAAGGCAA----CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCA ...............-.(((((((......(((((((....))))-))).(((((.(....)...((((----(.....))))).)))))((((((((.......))))))))))))))) ( -35.10) >DroEre_CAF1 66734 115 - 1 ACUUGGCUGUAAUUU-CAGGCCAUGGAGUUGAGAAGGGGAUUCUUUUUUGUGGGCAGUAAACAAGGCAA----CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCA ...(((((((((.((-(.(.(((..(((..(((((.....))))))))..))).)..........((((----(.....))))).((((((((.....)))))).))))).))))))))) ( -37.70) >DroYak_CAF1 64306 115 - 1 ACUUGGCUGUAAUUU-CUGGCCAUGGAAUUGCGAAGGGGAUUCUUUUUUGUGGGCAGUAAACAAGGCAA----CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCA ...(((((((((.((-(((.((((((((((.(....).)))))).....)))).)).........((((----(.....))))).((((((((.....)))))).))))).))))))))) ( -38.70) >consensus AGUUGGCUGUAAUUU_UUGGCCAUGGUAUUGAGAAGGGGAUCCUUUUUUGUGGGCAGUAAACAAGGCAA____CCCAAAGUUGCAGCUCGGAAUUCGCUUUCGAUGCGAAUUUAUGGCCA ....(((((((((((.(((.(((..(.....((((((....)))))))..))).))).......((........)).)))))))))))..((((((((.......))))))))....... (-30.36 = -30.36 + -0.00)

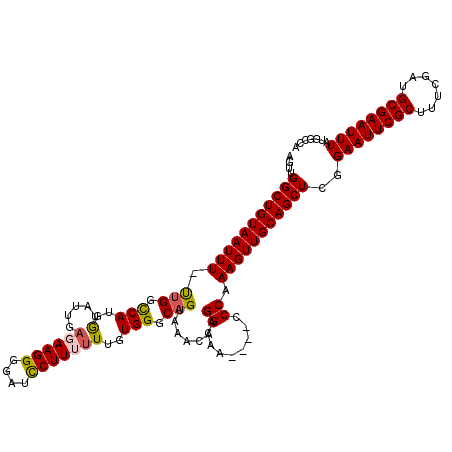

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:41 2006