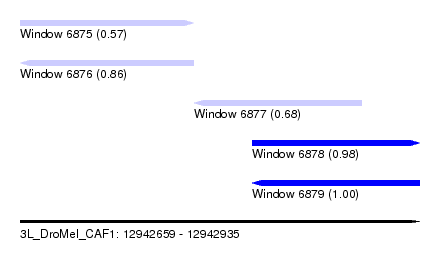
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,942,659 – 12,942,935 |
| Length | 276 |
| Max. P | 0.996969 |

| Location | 12,942,659 – 12,942,779 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -26.27 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12942659 120 + 23771897 GGAUAAACUGACUUAAAUUCAGUUGAAAUUGCCUUUGAAAGUAAAAUUUAUGAAGACAAACAAAAGCAGGAAAACUUGGCAGGACUCGCGGGCGAGUACCCGGCCAAUUGCCAAAGGACU .....((((((.......)))))).......((((((.............((....)).......((((......(((((.(((((((....))))).))..)))))))))))))))... ( -27.70) >DroSec_CAF1 59918 120 + 1 GGACAAACUGACUUAAAUUCAGUUGAAAUUGCCUUUGAAAGUAAAAUUUAUGAAGACAAACAAAAGCAGGAAAACUUGGCAGGACUCGCGGGCGAGUACCCGGCCAAUUGCCAAAGGACU .....((((((.......)))))).......((((((.............((....)).......((((......(((((.(((((((....))))).))..)))))))))))))))... ( -27.70) >DroEre_CAF1 66164 120 + 1 GAACUAACUGACUUAAAUUCAGUUGAAAUUCCCUUUGAAAGUAGAAUUUAUGAAGACAAACAAAAGCAGGCAAACUUGGCAGGACUCGCGGGCGAGUACCCGGCCAAUUGCCAAAGGACU ....(((((((.......)))))))......(((((..............((....))..........(((((..(((((.(((((((....))))).))..)))))))))))))))... ( -31.80) >DroYak_CAF1 63719 120 + 1 GAACAAACUGACUUAAAUUCAGUUGAAAUUUCCUUUGAACGUAAAAUUUAUGAAGACAAACAAAAGCAGGAAAACUUGGCAGGACUCGCGGGCGAGAACCCGGCCAAUUGCCAAAGGACU .....((((((.......))))))......(((((((..((((.....)))).............((((......(((((.((.((((....))))...)).)))))))))))))))).. ( -28.90) >consensus GAACAAACUGACUUAAAUUCAGUUGAAAUUGCCUUUGAAAGUAAAAUUUAUGAAGACAAACAAAAGCAGGAAAACUUGGCAGGACUCGCGGGCGAGUACCCGGCCAAUUGCCAAAGGACU .....((((((.......)))))).......((((((.............((....)).......((((......(((((.(((((((....))))).))..)))))))))))))))... (-26.27 = -26.52 + 0.25)



| Location | 12,942,659 – 12,942,779 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -27.95 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12942659 120 - 23771897 AGUCCUUUGGCAAUUGGCCGGGUACUCGCCCGCGAGUCCUGCCAAGUUUUCCUGCUUUUGUUUGUCUUCAUAAAUUUUACUUUCAAAGGCAAUUUCAACUGAAUUUAAGUCAGUUUAUCC .(((....)))..(((((.(((.(((((....))))))))))))).......((((((((...((.............))...)))))))).....((((((.......))))))..... ( -30.22) >DroSec_CAF1 59918 120 - 1 AGUCCUUUGGCAAUUGGCCGGGUACUCGCCCGCGAGUCCUGCCAAGUUUUCCUGCUUUUGUUUGUCUUCAUAAAUUUUACUUUCAAAGGCAAUUUCAACUGAAUUUAAGUCAGUUUGUCC ........((((.(((((.(((.(((((....))))))))))))).......((((((((...((.............))...)))))))).....((((((.......)))))))))). ( -31.52) >DroEre_CAF1 66164 120 - 1 AGUCCUUUGGCAAUUGGCCGGGUACUCGCCCGCGAGUCCUGCCAAGUUUGCCUGCUUUUGUUUGUCUUCAUAAAUUCUACUUUCAAAGGGAAUUUCAACUGAAUUUAAGUCAGUUAGUUC ..((((((((((((((((.(((.(((((....)))))))))))))..))))).......((((((....))))))..........)))))).....((((((.......))))))..... ( -34.50) >DroYak_CAF1 63719 120 - 1 AGUCCUUUGGCAAUUGGCCGGGUUCUCGCCCGCGAGUCCUGCCAAGUUUUCCUGCUUUUGUUUGUCUUCAUAAAUUUUACGUUCAAAGGAAAUUUCAACUGAAUUUAAGUCAGUUUGUUC ..(((((((((..(((((.(((..((((....)))).))))))))..............((((((....)))))).....).))))))))......((((((.......))))))..... ( -29.50) >consensus AGUCCUUUGGCAAUUGGCCGGGUACUCGCCCGCGAGUCCUGCCAAGUUUUCCUGCUUUUGUUUGUCUUCAUAAAUUUUACUUUCAAAGGCAAUUUCAACUGAAUUUAAGUCAGUUUGUCC ...(((((((((.(((((.(((.(((((....))))))))))))).......)))....((((((....))))))........)))))).......((((((.......))))))..... (-27.95 = -28.20 + 0.25)

| Location | 12,942,779 – 12,942,895 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -23.50 |
| Energy contribution | -25.34 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12942779 116 - 23771897 AUUUUUCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCAAAC----GAAAGGCGGAAGGAUAAUUCAAAUUGUCCAUUACGCGCAGCUGCCGCUCGAUUUG ............((((.(((((..(((((((....................((((((....----....)))))).(((((((....))))))).....).))))))..))).)).)))) ( -35.70) >DroSec_CAF1 60038 114 - 1 AAU--UCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCAAAC----GAAAGGCGGAAGGAUAAUUCAAAUUGUCCAUUACGCGCAGCUGCCGCUCGAUUUG ...--.......((((.(((((..(((((((....................((((((....----....)))))).(((((((....))))))).....).))))))..))).)).)))) ( -35.70) >DroSim_CAF1 64381 118 - 1 AAU--UCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCCACCACCUGAAAGGCGGAAGGAUAAUUCAAAUUGUCCANNNNNNNNNNNNNNNNNNNNNNNNN ...--....((.(((..(((((.....)))))................(((((((((............))))).)))).........))).)).......................... ( -22.30) >DroEre_CAF1 66284 114 - 1 AAU--UCGUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCAACG----GAACGACGGACGGAUAAUUCAAAUUGUCCAUUACGCGCAGCUGCCGCUCGAUUUG ...--..((((((.....)))((.(((((((.................(((((((....))----)).....))).(((((((....))))))).....).)))))))))))........ ( -30.80) >DroYak_CAF1 63839 114 - 1 AAU--UCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUUCGCCAACG----GGAAGACGGACGGAUAAUUCAAAUUGUCCAUUACGCGCAGCUGCCGCUCGAUUUG ...--.......((((.(((((..(((((((.................(((.((.((....----))..)).))).(((((((....))))))).....).))))))..))).)).)))) ( -29.20) >consensus AAU__UCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCAACC____GAAAGGCGGAAGGAUAAUUCAAAUUGUCCAUUACGCGCAGCUGCCGCUCGAUUUG ..........(((.....)))((.((((((.....................((((((............)))))).(((((((....))))))).......))))))))........... (-23.50 = -25.34 + 1.84)


| Location | 12,942,819 – 12,942,935 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -42.76 |
| Consensus MFE | -37.68 |
| Energy contribution | -37.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12942819 116 + 23771897 AUCCUUCCGCCUUUC----GUUUGGCGGAGGAGUGUGAGUAAUAGUUUUGGCAGCCAGCGCCAUUUUGGCCAGGAAAAAUUAUGGAGAUCCAUGAGCCAAGGGCCACUACAUCGACUCCG .....((((((....----....))))))(((((.((((((.......((((.......))))...(((((.((.....((((((....)))))).))...))))).))).)))))))). ( -44.00) >DroSec_CAF1 60078 114 + 1 AUCCUUCCGCCUUUC----GUUUGGCGGAGGAGUGUGAGUAAUAGUUUUGGCAGCCAGCGCCAUUUUGGCCAGGA--AUUUAUGGAGAUCCAUGAGCCAAGGGCCACUACAUCGACUCCG .....((((((....----....))))))(((((.((((((.......((((.......))))...(((((.((.--..((((((....)))))).))...))))).))).)))))))). ( -44.20) >DroSim_CAF1 64421 118 + 1 AUCCUUCCGCCUUUCAGGUGGUGGGCGGAGGAGUGUGAGUAAUAGUUUUGGCAGCCAGCGCCAUUUUGGCCAGGA--AUUUAUGGAGAUCCAUGAGCCAAGGGCCACUACAUCGACUCCG .(((.((((((........)))))).)))(((((.((((((.......((((.......))))...(((((.((.--..((((((....)))))).))...))))).))).)))))))). ( -46.10) >DroEre_CAF1 66324 114 + 1 AUCCGUCCGUCGUUC----CGUUGGCGGAGGAGUGUGAGUAAUAGUUUUGGCAGCCAGCGCCAUUUUGGCCACGA--AUUUAUGGAGAUCCAUGAGCCAAGGGCCACUACAUCGCCUCCG .....(((((((...----...)))))))((((.(((((((.......((((.......))))...(((((....--.(((((((....))))))).....))))).))).)))))))). ( -43.60) >DroYak_CAF1 63879 114 + 1 AUCCGUCCGUCUUCC----CGUUGGCGAAGGAGUGUGAGUAAUAGUUUUGGCAGCCAGCGCCAUUUUGGCCAGGA--AUUUAUGGAGAUCCAUGGGCCAAGGGCCACUACAUCGACUCCG .......((((....----....))))..(((((.((((((.......((((.......))))...(((((.((.--..((((((....)))))).))...))))).))).)))))))). ( -35.90) >consensus AUCCUUCCGCCUUUC____GGUUGGCGGAGGAGUGUGAGUAAUAGUUUUGGCAGCCAGCGCCAUUUUGGCCAGGA__AUUUAUGGAGAUCCAUGAGCCAAGGGCCACUACAUCGACUCCG .....((((((............))))))(((((.((((((.......((((.......))))...(((((.((.....((((((....)))))).))...))))).))).)))))))). (-37.68 = -37.88 + 0.20)

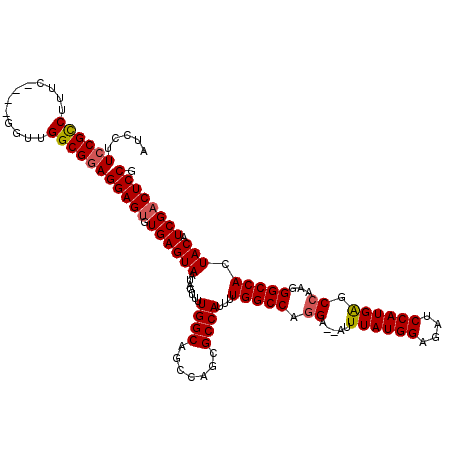
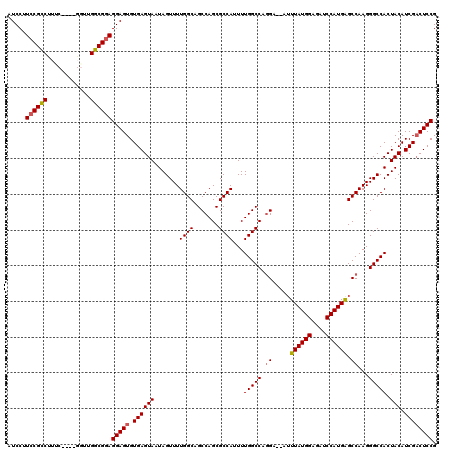
| Location | 12,942,819 – 12,942,935 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -39.76 |
| Consensus MFE | -35.52 |
| Energy contribution | -35.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12942819 116 - 23771897 CGGAGUCGAUGUAGUGGCCCUUGGCUCAUGGAUCUCCAUAAUUUUUCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCAAAC----GAAAGGCGGAAGGAU .(((((.((.(((((((...(((((..((((....)))).......((.((((.....))).).))..)))))..)))))))))..)))))((((((....----....))))))..... ( -41.50) >DroSec_CAF1 60078 114 - 1 CGGAGUCGAUGUAGUGGCCCUUGGCUCAUGGAUCUCCAUAAAU--UCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCAAAC----GAAAGGCGGAAGGAU .(((((.((.(((((((...(((((..((((....))))....--.((.((((.....))).).))..)))))..)))))))))..)))))((((((....----....))))))..... ( -41.50) >DroSim_CAF1 64421 118 - 1 CGGAGUCGAUGUAGUGGCCCUUGGCUCAUGGAUCUCCAUAAAU--UCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCCACCACCUGAAAGGCGGAAGGAU .(((((.((.(((((((...(((((..((((....))))....--.((.((((.....))).).))..)))))..)))))))))..)))))((((((............))))))..... ( -39.90) >DroEre_CAF1 66324 114 - 1 CGGAGGCGAUGUAGUGGCCCUUGGCUCAUGGAUCUCCAUAAAU--UCGUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCAACG----GAACGACGGACGGAU .(((((.((.(((((((...(((((.(((((((........))--))))))))))..(((((.....)))))...))))))))).).))))((((((..((----...))..)).)))). ( -40.50) >DroYak_CAF1 63879 114 - 1 CGGAGUCGAUGUAGUGGCCCUUGGCCCAUGGAUCUCCAUAAAU--UCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUUCGCCAACG----GGAAGACGGACGGAU .(((((.((.(((((((...((((((...(((...........--))).))))))..(((((.....)))))...)))))))))..)))))((((((..(.----....)..)).)))). ( -35.40) >consensus CGGAGUCGAUGUAGUGGCCCUUGGCUCAUGGAUCUCCAUAAAU__UCCUGGCCAAAAUGGCGCUGGCUGCCAAAACUAUUACUCACACUCCUCCGCCAACC____GAAAGGCGGAAGGAU .(((((....(((((((...(((((..((((....))))..........(((((.........))))))))))..)))))))....)))))((((((............))))))..... (-35.52 = -35.96 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:36 2006