
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,926,922 – 12,927,061 |
| Length | 139 |
| Max. P | 0.999854 |

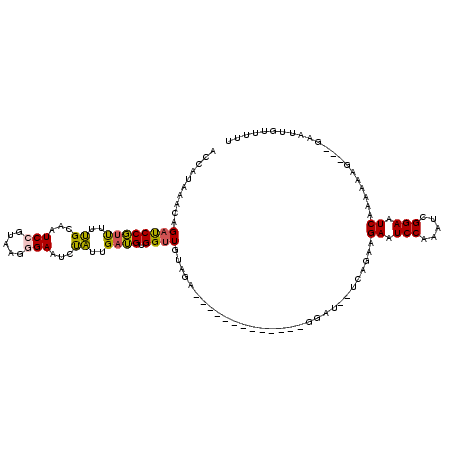
| Location | 12,926,922 – 12,927,021 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.91 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -10.92 |
| Energy contribution | -10.20 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12926922 99 + 23771897 AC------CAGAUCCGUUUUUGCAAUCCGUAAAGGAAUCUAUUGAUGUGGUUGUAGAGGA----------GGAU--UCAGAAGAAUCCAAAUCGGAAUCAAAAACG---GAAUGGUUUUU ((------((..((((((((((...((((........(((((..........)))))...----------((((--((....))))))....))))..))))))))---)).)))).... ( -31.90) >DroSec_CAF1 47648 102 + 1 AACAUAAACAGUUCCGUUUUUGCAAUCCGUCAGAGAAUCUAUUGAUGUGGUUGUAGA-------------GGAU--UCAGAAGAAUCCAAAUCGGAAUCAAAAAAG---GAAUUGUUUUU .....((((((((((...(((((((((((((((........)))))).)))))))))-------------((((--((....)))))).................)---))))))))).. ( -33.10) >DroSim_CAF1 51442 102 + 1 AACAUAAACAGAUCCGUUUUUGCUAUCCGUUAGAGAAUCUAUUGAUGUGGUUGUAGA-------------GGAU--UCAGAAGAAUCCAAAUCGGAAUCAAUAAAG---GAAUAGUUUUU ..........((((((..(((((.(((((((((........)))))).))).)))))-------------((((--((....))))))....))).))).......---........... ( -20.80) >DroEre_CAF1 53204 120 + 1 ACUGUGAACAGACCCUUCGUCGCAAUCCGUGAAGGAAUCCGUUGUGGAGGUUGUAGAGGAUUCCUGAGAUGGAUCAUCAGAAGAAUCCGAAUCGGAAUCAGUAUUCCACUUAACAGGCUU .((((.(((.((.((((((.((.....))))))))..)).)))((((((.........((((((.((..(((((..((....)))))))..))))))))....))))))...)))).... ( -33.32) >DroYak_CAF1 50771 111 + 1 ACUGUGCACAGACCCUUUGUCGCUGUCCGAAAGGGAAUCUGUUGAGGAGGUUGUAG------CCUGGGAUGGAUCACCAGAAGAAUCCGAAUCGGAAUCAGAAUCG---AUAUUGGGCUU .(((....))).(((..(((((((((((((...(((.(((.......((((....)------)))((((....)).))...))).)))...)))))..)))...))---)))..)))... ( -31.70) >consensus ACCAUAAACAGAUCCGUUUUUGCAAUCCGUAAGGGAAUCUAUUGAUGUGGUUGUAGA_____________GGAU__UCAGAAGAAUCCAAAUCGGAAUCAAAAAAG___GAAUUGUUUUU ..........((((((((..((...(((.....)))...))..)))).))))..............................((.(((.....))).))..................... (-10.92 = -10.20 + -0.72)


| Location | 12,926,922 – 12,927,021 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.91 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -9.74 |
| Energy contribution | -9.18 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.34 |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12926922 99 - 23771897 AAAAACCAUUC---CGUUUUUGAUUCCGAUUUGGAUUCUUCUGA--AUCC----------UCCUCUACAACCACAUCAAUAGAUUCCUUUACGGAUUGCAAAAACGGAUCUG------GU ....((((.((---((((((((..((((....((((((....))--))))----------...((((............))))........))))...))))))))))..))------)) ( -31.20) >DroSec_CAF1 47648 102 - 1 AAAAACAAUUC---CUUUUUUGAUUCCGAUUUGGAUUCUUCUGA--AUCC-------------UCUACAACCACAUCAAUAGAUUCUCUGACGGAUUGCAAAAACGGAACUGUUUAUGUU ..(((((.(((---(.((((((..((((....((((((....))--))))-------------................(((.....))).))))...)))))).)))).)))))..... ( -27.90) >DroSim_CAF1 51442 102 - 1 AAAAACUAUUC---CUUUAUUGAUUCCGAUUUGGAUUCUUCUGA--AUCC-------------UCUACAACCACAUCAAUAGAUUCUCUAACGGAUAGCAAAAACGGAUCUGUUUAUGUU ..((((...((---(.((.(((..((((....((((((....))--))))-------------................(((.....))).))))...))).)).)))...))))..... ( -22.10) >DroEre_CAF1 53204 120 - 1 AAGCCUGUUAAGUGGAAUACUGAUUCCGAUUCGGAUUCUUCUGAUGAUCCAUCUCAGGAAUCCUCUACAACCUCCACAACGGAUUCCUUCACGGAUUGCGACGAAGGGUCUGUUCACAGU ....((((...(((((.....((.......))((((((((..((((...))))..)))))))).........)))))(((((((.(((((.((.....))..)))))))))))).)))). ( -35.30) >DroYak_CAF1 50771 111 - 1 AAGCCCAAUAU---CGAUUCUGAUUCCGAUUCGGAUUCUUCUGGUGAUCCAUCCCAGG------CUACAACCUCCUCAACAGAUUCCCUUUCGGACAGCGACAAAGGGUCUGUGCACAGU .((((......---.((.(((((.......))))).))...(((.((....)))))))------))............(((((..((((((((.....)))..))))))))))....... ( -26.70) >consensus AAAAACAAUUC___CGUUUUUGAUUCCGAUUUGGAUUCUUCUGA__AUCC_____________UCUACAACCACAUCAAUAGAUUCCCUUACGGAUUGCAAAAACGGAUCUGUUCACAGU ....................((..((((....(((.(((..(((...............................)))..))).)))....))))...)).................... ( -9.74 = -9.18 + -0.56)



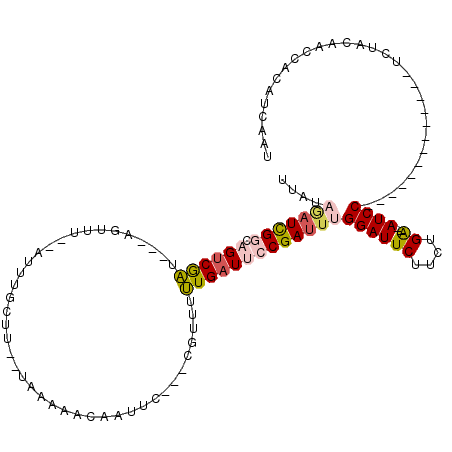
| Location | 12,926,956 – 12,927,061 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.48 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -7.28 |
| Energy contribution | -8.00 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12926956 105 - 23771897 UUAUACUUCGGCAGUCGAGUUGGAUUUGGAUUUGCUUGCUAAAAACCAUUC---CGUUUUUGAUUCCGAUUUGGAUUCUUCUGA--AUCC----------UCCUCUACAACCACAUCAAU .......((((.(((((((.((((..(((.(((........))).))).))---))..)))))))))))...((((((....))--))))----------.................... ( -26.00) >DroSec_CAF1 47688 99 - 1 UUAUAGAUCGGCAGUCGAU---AGUUUAUAUUUGCUUUAUAAAAACAAUUC---CUUUUUUGAUUCCGAUUUGGAUUCUUCUGA--AUCC-------------UCUACAACCACAUCAAU ....(((((((.((((((.---..((((((.......))))))........---.....)))))))))))))((((((....))--))))-------------................. ( -19.99) >DroSim_CAF1 51482 99 - 1 UUAUAGAUCGGCAGUCGAU---AGUUUAUAUUUGCUUUAUAAAAACUAUUC---CUUUAUUGAUUCCGAUUUGGAUUCUUCUGA--AUCC-------------UCUACAACCACAUCAAU ....(((((((.(((((((---((((((((.......))))))........---..))))))))))))))))((((((....))--))))-------------................. ( -22.70) >DroEre_CAF1 53244 108 - 1 UUAUAGAUCGUCUGUCAGU---AG-----AAUUGCU----AAGCCUGUUAAGUGGAAUACUGAUUCCGAUUCGGAUUCUUCUGAUGAUCCAUCUCAGGAAUCCUCUACAACCUCCACAAC .....(((((...((((((---(.-----..(..((----.((....)).))..)..)))))))..))))).((((((((..((((...))))..))))))))................. ( -27.50) >DroYak_CAF1 50811 99 - 1 UUAUAAAUUGUCUGCCGGU---GG-----AAUUGCU----AAGCCCAAUAU---CGAUUCUGAUUCCGAUUCGGAUUCUUCUGGUGAUCCAUCCCAGG------CUACAACCUCCUCAAC .........(((((..(((---((-----(.(..((----(.(........---)((.(((((.......))))).))...)))..))))))).))))------)............... ( -20.70) >consensus UUAUAGAUCGGCAGUCGAU___AGUUU__AUUUGCUU__UAAAAACAAUUC___CGUUUUUGAUUCCGAUUUGGAUUCUUCUGA__AUCC_____________UCUACAACCACAUCAAU ....(((((((.((((((.........................................)))))))))))))((((((....))..)))).............................. ( -7.28 = -8.00 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:25 2006