
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,920,953 – 12,921,062 |
| Length | 109 |
| Max. P | 0.619492 |

| Location | 12,920,953 – 12,921,062 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -23.11 |
| Consensus MFE | -12.10 |
| Energy contribution | -13.93 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12920953 109 + 23771897 CGUUGAAACAAAAACUUCAACGGCGGGCAAAAAAAU-AUAUACAUAUAUUUGGGGCGCGGAGCUGAGAAGGAAAAUCGGCCUAAUACCCAUAUUGAUAACAUAAGCCGAA (((((((........)))))))...(((....((((-(((....)))))))(((....((.(((((.........)))))))....)))...............)))... ( -26.60) >DroSec_CAF1 41673 106 + 1 CGUUGAAACAAAAACUUCAACGGCGGGCAAAAAA----UAUAUAUAUAUUUGGGGCGCGGAGCUGAGAAGGAAAAUCGGCCUAAUACCCAUAUUGAUAACAUAAGCCAAA (((((((........)))))))...(((...(((----(((....))))))(((....((.(((((.........)))))))....)))...............)))... ( -25.50) >DroSim_CAF1 40760 110 + 1 CGUUGAAACAAAAACUUCAACGGCGGGCAAAAAAAAAGUAUAUAUAUAUUUGGGGCGCGGAGCUGAGAAGGAAAAUCGGCCUAAUACCCAUAUUGAUAACAUAAGCCAAA (((((((........)))))))...(((.......((((((....))))))(((....((.(((((.........)))))))....)))...............)))... ( -25.50) >DroEre_CAF1 47088 99 + 1 CGUUGAAACAAAAACUUCAACGGCGGAGAAAAAAAU-AU--------AUGUGGG--GCGGAGCUGAGCAGGAAAAUCACCCUAAUACCCAUAUUGAUAACUUAAGCCAAA .((((((........))))))(((............-..--------(((((((--((........))(((........)))....)))))))...........)))... ( -23.11) >DroYak_CAF1 44680 103 + 1 CGUUCAAACAAAAACUUCAACGGCGGAGAAAAAAAA-AU----AUAAAUGUGGG--GUGAAGCUGAGAGGGAAAAUCAGCCUAAUACCCAUAUUGAUAACUUAAGCCAAA .....................(((.(((........-.(----((.((((((((--.....(((((.........)))))......)))))))).))).)))..)))... ( -19.90) >DroPer_CAF1 47176 92 + 1 CGUUGAAACGAAAACUUCAACGGAUGGAGAAAAA----UGUACGU----------CGUGGGG----AAAGAGAAAGCAACCUAAUACCCAUAUUGAUAACUUAAGCCAGA (((((((........)))))))..(((.......----(((.((.----------.(((((.----....((........))....)))))..))))).......))).. ( -18.04) >consensus CGUUGAAACAAAAACUUCAACGGCGGGCAAAAAAA__AUAUA_AUAUAUUUGGG_CGCGGAGCUGAGAAGGAAAAUCAGCCUAAUACCCAUAUUGAUAACAUAAGCCAAA .((((((........))))))(((..........................((((....((.(((((.........)))))))....))))..(((......))))))... (-12.10 = -13.93 + 1.83)

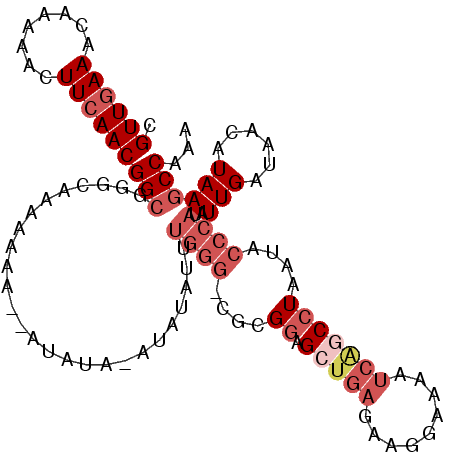
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:21 2006