| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,910,276 – 12,910,367 |
| Length | 91 |
| Max. P | 0.791218 |

| Location | 12,910,276 – 12,910,367 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
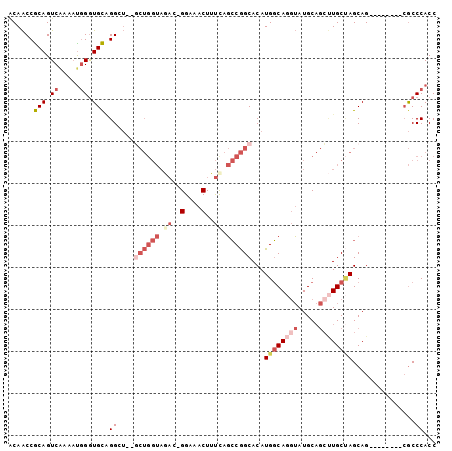
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -35.68 |
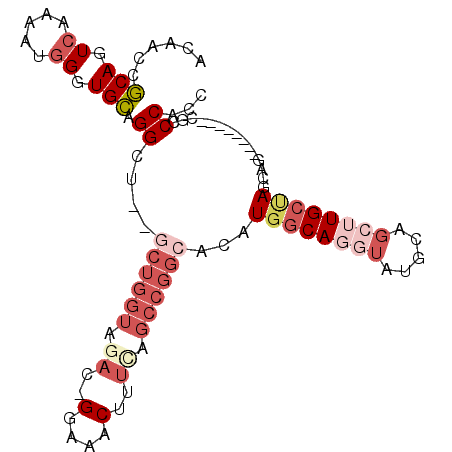
| Consensus MFE | -17.19 |
| Energy contribution | -19.72 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12910276 91 + 23771897 ACAACCGCAGUCAAAAUGGGUGCUGGCU--GCUGGUAGAC-GGAAACUUUCAGCCGGCACAUGGCAGGUAUGCAGCUUGCUAGCAG--------CGCCCACC ................(((((((((..(--((((((.((.-(....)..)).)))))))..((((((((.....)))))))).)))--------)))))).. ( -39.70) >DroPse_CAF1 39957 83 + 1 GCAGCCACAGUCGAACUCGAUGUGGG---------------GGAAACU-CCAUGCAGCACAUGCCAU-AAUGC--CCUGCCAGCAGGUAUGUGGGACCCACC ((.......((((....))))(((((---------------(....))-))))))..((((((((..-..(((--.......)))))))))))......... ( -28.30) >DroSec_CAF1 31041 91 + 1 ACAACCGCAGUCAAAAUGGGUGCAGGCU--GCUGGUAGAC-GGAAACUUUCAGCCGGCACAUGGCAGGUAUGCAGCUUGCAAGCAG--------CGCCCACC ................(((((((..(((--((((((.((.-(....)..)).)))))).....((((((.....)))))).))).)--------)))))).. ( -35.90) >DroSim_CAF1 28855 91 + 1 ACAACCGCAGUCAAAAUGGGUGCAGGCU--GCUGGUAGAC-GGAAACUUUCAGCCGGCACAUGGCAGGUAUGCAGCUUGCUAGCAG--------CGCCCACC ................(((((((..(((--((((((.((.-(....)..)).)))))))..((((((((.....)))))))))).)--------)))))).. ( -37.20) >DroEre_CAF1 36358 91 + 1 ACAACCGCAGUCAAAACGGGUGCAGGAU--UCUGGUAGAG-GGAAACUUUUAGCCGGCACAUGGCAGGUAUGCAGAUUGCUAGCAG--------CGCCCACC .................((((((.....--.....(((((-(....))))))((..(((....(((....)))....)))..)).)--------)))))... ( -30.80) >DroYak_CAF1 33546 94 + 1 ACAACCGCAGUCAAAAUGGGUGCAGGAUGUGCUGGUAAAGGGGAAACUUUCAGCCGGCACAUGGCAGGUAUGCAGCAUGCUAGCAG--------CGCCCACC ................(((((((...((((((((((..((((....))))..)))))))))).((.((((((...)))))).)).)--------)))))).. ( -42.20) >consensus ACAACCGCAGUCAAAAUGGGUGCAGGCU__GCUGGUAGAC_GGAAACUUUCAGCCGGCACAUGGCAGGUAUGCAGCUUGCUAGCAG________CGCCCACC ......(((.((.....)).))).((....((((((.((..(....)..)).))))))...((((((((.....))))))))...............))... (-17.19 = -19.72 + 2.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:08 2006