
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,909,266 – 12,909,407 |
| Length | 141 |
| Max. P | 0.972583 |

| Location | 12,909,266 – 12,909,375 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.03 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -26.74 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
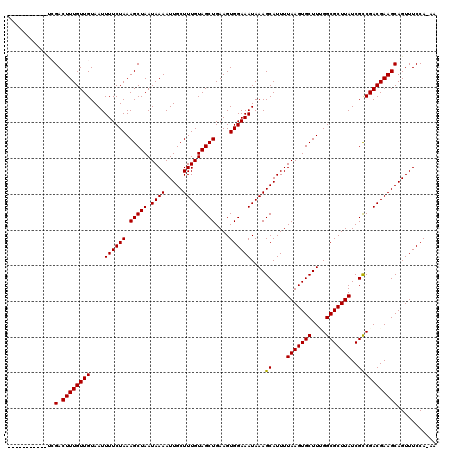
>3L_DroMel_CAF1 12909266 109 + 23771897 -----------UCGACUUUGUUGUAAUUUUCUAAAGCUAAUAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUGGGCGCUUAUCGCCGACGAAGCAGUUUCCAAAA -----------..(.((((((((....((((((.(((((.((((.....)))))))))....))))))....((....(((((((....)))))))..)))))))))))........... ( -27.00) >DroPse_CAF1 39035 116 + 1 CGUCGACG---CAGACUUUGUUGUAAUUUUCUAAAGCUAAUAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUUGGCGCUUAUCGCCGACGAAGCAGUUUCCA-AA .(((....---..)))..........................((((((((((((.((..(((((....((((((((.....)))))))).)))))...))..))))))))))))...-.. ( -29.90) >DroSec_CAF1 30036 109 + 1 -----------UCGACUUUGUUGUAAUUUUCUAAAGCUAAUAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUCGGCGCUUAUCGCCGACGAAGCAGUUUCCAAAA -----------..(.((((((((....((((((.(((((.((((.....)))))))))....))))))....((....(((((((....)))))))..)))))))))))........... ( -30.00) >DroYak_CAF1 32570 109 + 1 -----------UCGACUUUGUUGUAAUUUUCUAAAGCUAAUAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUGGGCGCUUAUCGCCGACGAAGCAGUUUCCAAAA -----------..(.((((((((....((((((.(((((.((((.....)))))))))....))))))....((....(((((((....)))))))..)))))))))))........... ( -27.00) >DroAna_CAF1 56759 119 + 1 UGUCCUGGCAGGCGACUUUGUUGUAAUUUUCUAAAGCUAAUAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUUGGCGCUUAUCGUCGACGAAGCAGUUUCCA-AA .....(((.((((..((((((((.......(((((((...((((((.(((((((..((.....))..)))))))))))))...)))))))..........))))))))..)))))))-.. ( -33.13) >DroPer_CAF1 34656 116 + 1 CGUCGACG---CAGACUUUGUUGUAAUUUUCUAAAGCUAAUAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUUGGCGCUUAUCGCCGACGAAGCAGUUUCCA-AA .(((....---..)))..........................((((((((((((.((..(((((....((((((((.....)))))))).)))))...))..))))))))))))...-.. ( -29.90) >consensus ___________UCGACUUUGUUGUAAUUUUCUAAAGCUAAUAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUUGGCGCUUAUCGCCGACGAAGCAGUUUCCA_AA .............(.((((((((....((((((.(((((.((((.....)))))))))....))))))....((....(((((((....)))))))..)))))))))))........... (-26.74 = -27.10 + 0.36)


| Location | 12,909,295 – 12,909,407 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.32 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.40 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
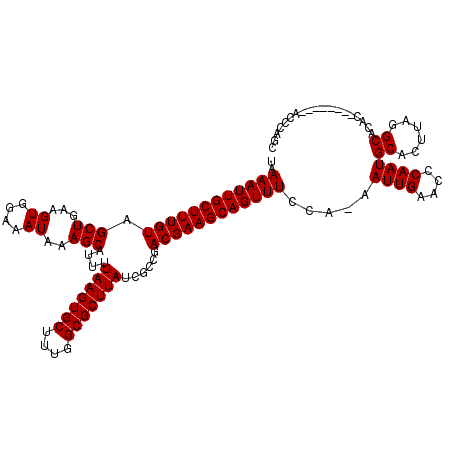
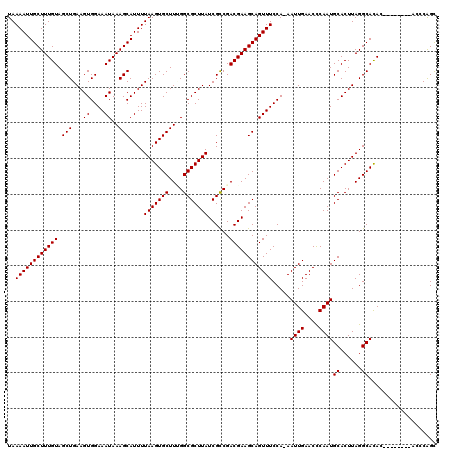
>3L_DroMel_CAF1 12909295 112 + 23771897 UAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUGGGCGCUUAUCGCCGACGAAGCAGUUUCCAAAAUUGAACCCAAUGCACUUAGGCACAC--------ACCCAGC ......((((((((..((.....))..)))))))).((((((((((((((.(((..(((....))))))...(((.......)))))))).))))))))).....--------....... ( -28.50) >DroPse_CAF1 39072 115 + 1 UAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUUGGCGCUUAUCGCCGACGAAGCAGUUUCCA-AAUUGAACUCAAUGCACUUAGGCACAGAAAAG----CACAGU ......((((((((.(((.(((((((((((((((((.....)))))))((((.....)))).........)))))).-.((((....))))..)))).))))))...)))----)).... ( -31.50) >DroSec_CAF1 30065 112 + 1 UAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUCGGCGCUUAUCGCCGACGAAGCAGUUUCCAAAAUUGAACCCAAUGCACUUAGGCACAC--------ACCCAGC ..((((((((((((.(((...((....))..)))....(((((((....)))))))......))))))))))))................(((......)))...--------....... ( -29.50) >DroYak_CAF1 32599 112 + 1 UAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUGGGCGCUUAUCGCCGACGAAGCAGUUUCCAAAAUUGAACCCAAUGCACUUAGGCACAC--------ACCCAGC ......((((((((..((.....))..)))))))).((((((((((((((.(((..(((....))))))...(((.......)))))))).))))))))).....--------....... ( -28.50) >DroAna_CAF1 56799 119 + 1 UAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUUGGCGCUUAUCGUCGACGAAGCAGUUUCCA-AAUUGAACUCAAUGCACUUAGGCAUAGGAAACUCCGGUCGGC ......((((((((..((.....))..))))))))...(((((((....)))))))..((((((...(.(((((((.-...((....))((((......)))).))))))).).)))))) ( -36.60) >DroPer_CAF1 34693 115 + 1 UAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUUGGCGCUUAUCGCCGACGAAGCAGUUUCCA-AAUUGAACUCAAUGCACUUAGGCACAGAAAAG----CACAGU ......((((((((.(((.(((((((((((((((((.....)))))))((((.....)))).........)))))).-.((((....))))..)))).))))))...)))----)).... ( -31.50) >consensus UAAAAUUGCUUUGUAGCUGAAGUGGAAAUAAAGCAUUUUAAGUGCUUUGGCGCUUAUCGCCGACGAAGCAGUUUCCA_AAUUGAACCCAAUGCACUUAGGCACAC________ACCCAGC ..((((((((((((.((..(((((......((((((.....))))))...)))))...))..)))))))))))).....((((....))))((......))................... (-25.54 = -25.40 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:06 2006