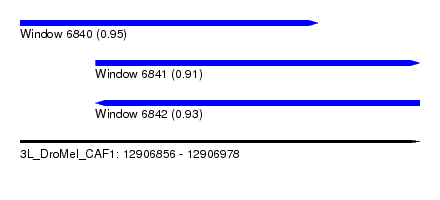
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,906,856 – 12,906,978 |
| Length | 122 |
| Max. P | 0.950664 |

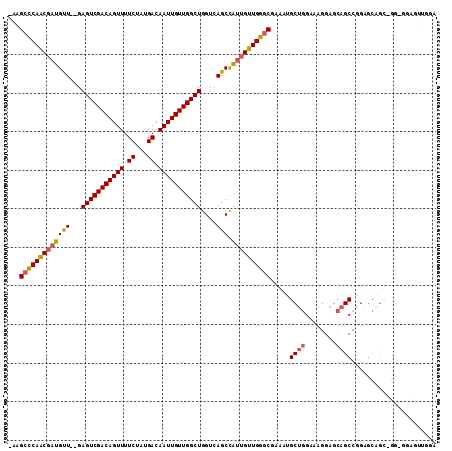
| Location | 12,906,856 – 12,906,947 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -28.17 |
| Energy contribution | -27.98 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12906856 91 + 23771897 -----CGACCU-----------UUCAACCCUCUGACUUG-AAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAA -----......-----------(((((.........)))-))(((((((((((((--((((((((((((.((...)).))))))))))...)))).)))))))))))... ( -32.60) >DroSec_CAF1 27648 91 + 1 -----CGACCU-----------UUCAACCCUCUGACUUG-AAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCAAA -----......-----------(((((.........)))-))(((((((((((((--((((((((((((.((...)).))))))))))...)))).)))))))))))... ( -32.60) >DroSim_CAF1 25437 91 + 1 -----CGACCU-----------UUCGACCCUCUGACUCG-AAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAA -----......-----------(((((.........)))-))(((((((((((((--((((((((((((.((...)).))))))))))...)))).)))))))))))... ( -34.40) >DroEre_CAF1 32859 94 + 1 ---GACGACCU-----------UUCAACCCUCUGACUGCCAAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAA ---........-----------....................(((((((((((((--((((((((((((.((...)).))))))))))...)))).)))))))))))... ( -32.00) >DroYak_CAF1 30232 91 + 1 -----CGACCU-----------UUCAACCCUCUGACUGG-AAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAA -----...((.-----------.(((......)))..))-..(((((((((((((--((((((((((((.((...)).))))))))))...)))).)))))))))))... ( -35.60) >DroAna_CAF1 54550 107 + 1 UCAGGCGACCCCGUCCUACGCCAUCGACC--CUGACAUA-GAGCUCAGCCCGGCUAGGAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCUUUUGUUGGCCCAA ..(((((....)).)))..((((.(((..--(((((...-.((((......))))...(((((((((((.((...)).))))))))))).)))))...))).)))).... ( -32.80) >consensus _____CGACCU___________UUCAACCCUCUGACUUG_AAGCCCAACGAUGUU__GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAA ..........................................(((((((((((((...(((((((((((.((...)).)))))))))))....))).))))))))))... (-28.17 = -27.98 + -0.19)

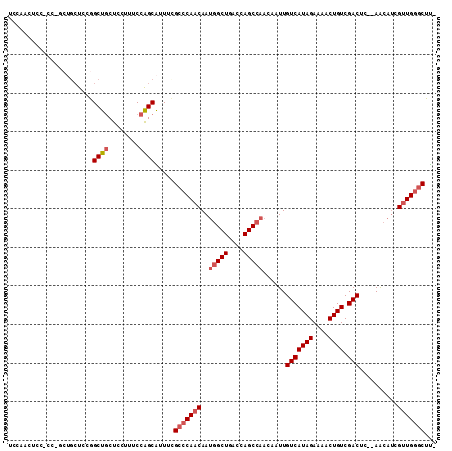
| Location | 12,906,879 – 12,906,978 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -31.27 |
| Energy contribution | -31.42 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12906879 99 + 23771897 -AAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAAUCGCUGGAAAGGAGCAGCCGGA---------GGAGUUGGA -..(((((((((((((--((((((((((((.((...)).))))))))))...)))).))))))))))).....((((........))))....---------......... ( -36.50) >DroSec_CAF1 27671 108 + 1 -AAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCAAAAUGCUGGAAAGGAGCAGCCGGAGCAGCCGGUGGAGUUGGA -..(((((((((((((--((((((((((((.((...)).))))))))))...)))).)))))))))))....((((.......))))(((((.....)))))......... ( -42.70) >DroSim_CAF1 25460 108 + 1 -AAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAAAUGCUGGAAAGGAGCAGCCGGAGCAGCCGGUGGAGUUGGA -..(((((((((((((--((((((((((((.((...)).))))))))))...)))).)))))))))))....((((.......))))(((((.....)))))......... ( -42.70) >DroEre_CAF1 32884 109 + 1 CAAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAAAUGCUGGAAAGGAGCAGCCGGAGCAGUUGGAGCAGUUGGA ...(((((((((((((--((((((((((((.((...)).))))))))))...)))).)))))))))))....((((.......)))).((((.((.......))..)))). ( -38.10) >DroYak_CAF1 30255 105 + 1 -AAGCCCAACGAUGUU--GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAACUGCUGGAAAGGAGCAGCCCGAGCAGUUGGAGCAG---GA -..(((((((((((((--((((((((((((.((...)).))))))))))...)))).))))))))))).(((((((.(...((.....))).))))))).......---.. ( -42.20) >DroAna_CAF1 54587 96 + 1 -GAGCUCAGCCCGGCUAGGAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCUUUUGUUGGCCCAAACGCCGGCGAGAAGGAGCUG-AGCUGC------------- -.((((((((((((((...(((((((((((.((...)).)))))))))))))))...((((((((((......)))))))))).)).))))-))))..------------- ( -44.80) >consensus _AAGCCCAACGAUGUU__GAGUCGACAGUUUUCUAUGACAAUUGUUGGCUGGUCAGCCAUUGUUGGGCGAAAUGCUGGAAAGGAGCAGCCGGAGCAGC_GG_GGAGUUGGA ...(((((((((((((...(((((((((((.((...)).)))))))))))....))).)))))))))).....((((........))))...................... (-31.27 = -31.42 + 0.14)

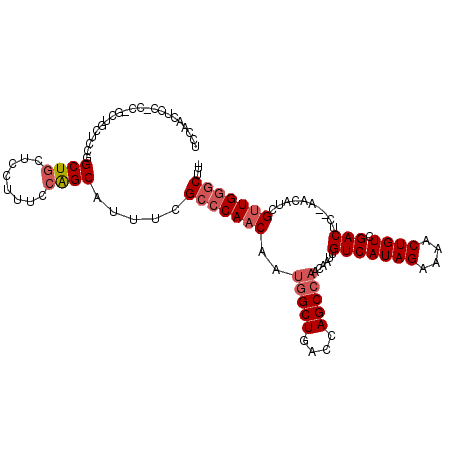
| Location | 12,906,879 – 12,906,978 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -24.99 |
| Energy contribution | -25.85 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12906879 99 - 23771897 UCCAACUCC---------UCCGGCUGCUCCUUUCCAGCGAUUCGCCCAACAAUGGCUGACCAGCCAACAAUUGUCAUAGAAAACUGUCGACUC--AACAUCGUUGGGCUU- .........---------....((((........)))).....(((((((..(((((....)))))......(((((((....)))).)))..--......)))))))..- ( -29.70) >DroSec_CAF1 27671 108 - 1 UCCAACUCCACCGGCUGCUCCGGCUGCUCCUUUCCAGCAUUUUGCCCAACAAUGGCUGACCAGCCAACAAUUGUCAUAGAAAACUGUCGACUC--AACAUCGUUGGGCUU- ..........((((.....)))).((((.......))))....(((((((..(((((....)))))......(((((((....)))).)))..--......)))))))..- ( -31.80) >DroSim_CAF1 25460 108 - 1 UCCAACUCCACCGGCUGCUCCGGCUGCUCCUUUCCAGCAUUUCGCCCAACAAUGGCUGACCAGCCAACAAUUGUCAUAGAAAACUGUCGACUC--AACAUCGUUGGGCUU- ..........((((.....)))).((((.......))))....(((((((..(((((....)))))......(((((((....)))).)))..--......)))))))..- ( -32.10) >DroEre_CAF1 32884 109 - 1 UCCAACUGCUCCAACUGCUCCGGCUGCUCCUUUCCAGCAUUUCGCCCAACAAUGGCUGACCAGCCAACAAUUGUCAUAGAAAACUGUCGACUC--AACAUCGUUGGGCUUG .......((.......))....((((........)))).....(((((((..(((((....)))))......(((((((....)))).)))..--......)))))))... ( -30.40) >DroYak_CAF1 30255 105 - 1 UC---CUGCUCCAACUGCUCGGGCUGCUCCUUUCCAGCAGUUCGCCCAACAAUGGCUGACCAGCCAACAAUUGUCAUAGAAAACUGUCGACUC--AACAUCGUUGGGCUU- ..---..((.......))..((((((((.......))))))))(((((((..(((((....)))))......(((((((....)))).)))..--......)))))))..- ( -36.40) >DroAna_CAF1 54587 96 - 1 -------------GCAGCU-CAGCUCCUUCUCGCCGGCGUUUGGGCCAACAAAAGCUGACCAGCCAACAAUUGUCAUAGAAAACUGUCGACUCCUAGCCGGGCUGAGCUC- -------------..((((-((((.........(((((....(((.........(((....)))........(((((((....)))).))))))..))))))))))))).- ( -30.40) >consensus UCCAACUCC_CC_GCUGCUCCGGCUGCUCCUUUCCAGCAUUUCGCCCAACAAUGGCUGACCAGCCAACAAUUGUCAUAGAAAACUGUCGACUC__AACAUCGUUGGGCUU_ ......................((((........)))).....(((((((..(((((....)))))......(((((((....)))).)))..........)))))))... (-24.99 = -25.85 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:01 2006