| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,902,390 – 12,902,496 |
| Length | 106 |
| Max. P | 0.577338 |

| Location | 12,902,390 – 12,902,496 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -19.45 |
| Consensus MFE | -13.43 |
| Energy contribution | -13.10 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
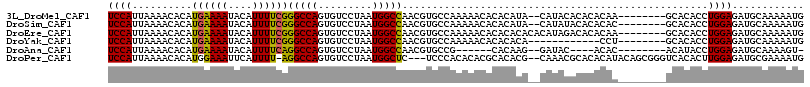
>3L_DroMel_CAF1 12902390 106 + 23771897 UCCAUUAAAACACAUGAAAAUACAUUUUCGGGCCAGUGUCCUAAUGGCCAACGUGCCAAAAACACACAUA--CAUACACACACAA--------GCACACCUGGAGAUGCAAAAAUG ......................(((((((((((((.........)))))...((((..............--.............--------))))...))))))))........ ( -17.63) >DroSim_CAF1 21142 106 + 1 UCCAUUAAAACACAUGAAAAUACAUUUUCGGGCCAGUGUCCUAAUGGCCAACGUGCCAAAAACACACAUA--CAUAUACACACAC--------GCACACCUGGAGAUGCAAAAAUG ......................((((((((((...((((.....((((......)))).....))))...--......(......--------)....))))))))))........ ( -17.60) >DroEre_CAF1 28512 108 + 1 UCCAUUAAAACACAUGAAAAUACAUUUUCGGGCCAGUGUCCUAAUGGCCAACGUGCCAAAAACACACACACACAUAGACACACAA--------GCACACCUGGAGAUGCAAAAAUG ......................((((((((((...((((.....((((......)))).....)))).........(.(......--------))...))))))))))........ ( -18.20) >DroYak_CAF1 25758 96 + 1 UCCAUUAAAACACAUGAAAAUACAUUUUCGGGCCAGUGUCCUAAUGGCCAACGUGCCAAAAACACACACA------------CCU--------GCACACCUGGAGAUGCAAAAAUG ......................(((((((((((((.........)))))...((((..............------------...--------))))...))))))))........ ( -17.93) >DroAna_CAF1 50255 95 + 1 UCCAUUAAAACACAUGAAAAUACAUUUUCAGGCCAGUGUCCUAAUGGCCAACGUGCCG------CACAAG--GAUAC----ACAC--------ACAUACCUGGAGAUGCAAAAGU- ......................((((((((((...(((((((..((((......))))------....))--)))))----....--------.....)))))))))).......- ( -22.10) >DroPer_CAF1 27782 110 + 1 UCCAUUAAAACACAUGGAAAUUCAUUUU-AGGCCAGUGUCCUAAUGGCUC---UCCCACACACGCACACG--CAAACGCACACAUACAGCGGGUCACACUUGGAGAUGCGAAAAUG ..(((((..((((.(((...........-...)))))))..)))))((((---(((.......((....)--)...(((.........)))..........))))).))....... ( -23.24) >consensus UCCAUUAAAACACAUGAAAAUACAUUUUCGGGCCAGUGUCCUAAUGGCCAACGUGCCAAAAACACACACA__CAUACACACACAA________GCACACCUGGAGAUGCAAAAAUG ((((..........((((((....))))))(((((.........)))))...................................................))))............ (-13.43 = -13.10 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:52 2006