| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,899,906 – 12,900,009 |
| Length | 103 |
| Max. P | 0.742801 |

| Location | 12,899,906 – 12,900,009 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -17.84 |
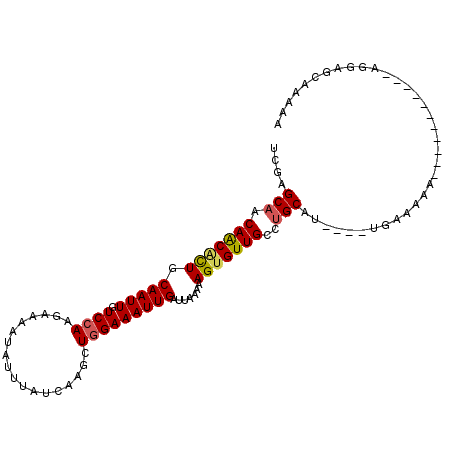
| Consensus MFE | -12.59 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12899906 103 + 23771897 UCGGGCAACAACACUGCAAUUGUCCAAGAAAAUAUUUAUCAAGCUGGAAAUUGAUUAAAAAGUGUUGCCUGCAC----UAAAAAAAAAAAAAGCAAAAGAGCAAGAG .(((((((((......(((((.((((..................))))))))).........)))))))))...----..............((......))..... ( -19.73) >DroSec_CAF1 20886 98 + 1 UCGGGCAACAACACUGCAAUUGUCCAAGAAAAUAUUUAUCAAGCUGGAAAUUGAUUAAAAAGUGUUGCCUGCAU----UGAAAAA-----AAGCAAAAGAGCAAAAG .(((((((((......(((((.((((..................))))))))).........)))))))))...----.......-----..((......))..... ( -19.73) >DroSim_CAF1 18723 88 + 1 UCGGGCAACAACACUGCAAUUGUCCAAGAAAAUAUUUAUCAAGCUGGAAAUUGAUUAAAAAGUGUUGCCUGCAU----UGAAAAAA----AAGCAA----------- .(((((((((......(((((.((((..................))))))))).........)))))))))...----........----......----------- ( -18.33) >DroEre_CAF1 26152 92 + 1 UCGAGCAACAACGCUGCAAUUGUCCAAGAAAAUAUUUAUCAAGCUAGAAAUUGAUUAAAAAGUGUUGCCUGCAU----UGGAAAA-----------AGGAGCACAAA ............(((.(.....(((((...((((((((((((........)))))....))))))).......)----))))...-----------.).)))..... ( -14.60) >DroYak_CAF1 23175 92 + 1 UCGAGCAGCAGCACUGCAAUUGUCCAAGAAAAUAUUUAUCAAGCUGGAAAUUGAUUAAAAAGUGUUGCCUGCAU----UGGAAAA-----------UGGAGCAAAAA (((((((((((((((.(((((.((((..................))))))))).......))))))).)))).)----)))....-----------........... ( -21.07) >DroAna_CAF1 48274 96 + 1 UUGAGCAACAACUUUGCAAUUGUCCAAGAAAAUAUUUAUCAAGCUGGAAAUUGAUUAAAAAGUGUUGGCAGCAAAAAGUGGAAAA-----------AGAAAUUCUAA (((..(((((.((((.(((((.((((..................))))))))).....))))))))).))).......(((((..-----------.....))))). ( -13.57) >consensus UCGAGCAACAACACUGCAAUUGUCCAAGAAAAUAUUUAUCAAGCUGGAAAUUGAUUAAAAAGUGUUGCCUGCAU____UGAAAAA___________AGGAGCAAAAA ....(((.(((((((.(((((.((((..................))))))))).......)))))))..)))................................... (-12.59 = -12.70 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:51 2006