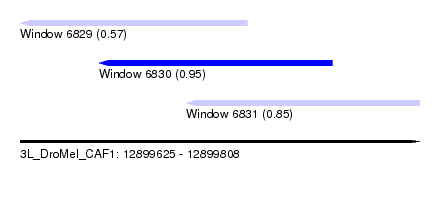
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,899,625 – 12,899,808 |
| Length | 183 |
| Max. P | 0.954008 |

| Location | 12,899,625 – 12,899,729 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -12.81 |
| Energy contribution | -13.28 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12899625 104 - 23771897 UCC------------ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUUCUCCAACGCACAUUGUCGCAAAAUAUGACAGUUGGU----UGGGAUGGCAAAGCGUAUAUCACAAUCGC ...------------...........(((((((((((.......((............))..(((((((.......))))))).)))----)))).))))...(((...........))) ( -23.30) >DroSec_CAF1 20605 104 - 1 UCC------------ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUUCUCCAACGCACAUUGUGGCAAAAUAUGACAGUUCGU----UGGGAUGGCAAAGCGUAUAUCACAAUCCC ...------------...........((((((((((.(((((((((............))....)).))))).....((....))))----)))).)))).................... ( -19.80) >DroSim_CAF1 18438 108 - 1 UCC------------ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUUCUCCAACGCACAUUGUCGCAAAAUAUGACAGUUCGUUGGUUGGGAUGGCAAAGCGUAUAUCACAAUCGC ...------------...........(((((((((....((...)).......((((((...(((((((.......))))))).))))))))))).))))...(((...........))) ( -26.70) >DroEre_CAF1 25869 100 - 1 UCC------------ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUCUUCUCCAAGGCACAUUGUCGCAAAAUAUGACAGU--------UGGGAUGGCACAGCGUAUAUCACAAUCGC ...------------...........((((((((((..((((...............))))...(((((.......)))))))--------)))).))))...(((...........))) ( -22.16) >DroYak_CAF1 22887 112 - 1 UCCACUUCCACAUCCACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUACUCCAACGCACAUUGUCGCAAAAUAUGACAGU--------UGGGAUGGCAAAGCGUAUAUCACCAUCAC ..........................((((((((((........((............))....(((((.......)))))))--------)))).)))).................... ( -18.20) >DroPer_CAF1 24042 83 - 1 U-C------------ACGUUACCCCACCCCACUA------------UUAUACCCCG----ACAUUGUCACAAAAGAUGACAGU--------UGGGCUGGCAAAACGUAUAUCACAAUGUC .-.------------(((((...(((.((((...------------..........----..((((((((....).)))))))--------)))).)))...)))))............. ( -17.26) >consensus UCC____________ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUUCUCCAACGCACAUUGUCGCAAAAUAUGACAGU________UGGGAUGGCAAAGCGUAUAUCACAAUCGC ..........................((((((((..........((............))..(((((((.......)))))))........)))).)))).................... (-12.81 = -13.28 + 0.47)

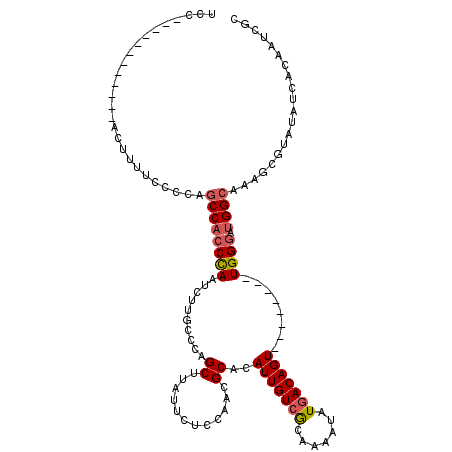
| Location | 12,899,661 – 12,899,768 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12899661 107 - 23771897 UGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC------------ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUUCUCCAACGCACAUUGUCGCAAAAUAUGAC .(((((((...)))))))((.(((.....)))..........------------.................((((..(((.................))).))))..)).......... ( -18.33) >DroSec_CAF1 20641 107 - 1 UGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC------------ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUUCUCCAACGCACAUUGUGGCAAAAUAUGAC .(((((((...)))))))...(((.....)))..........------------...........(((((.......(((.................)))....))))).......... ( -22.33) >DroSim_CAF1 18478 107 - 1 UGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC------------ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUUCUCCAACGCACAUUGUCGCAAAAUAUGAC .(((((((...)))))))((.(((.....)))..........------------.................((((..(((.................))).))))..)).......... ( -18.33) >DroEre_CAF1 25901 107 - 1 UGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC------------ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUCUUCUCCAAGGCACAUUGUCGCAAAAUAUGAC .(((((((...)))))))((.(((.....)))..........------------.................((((..((((...............)))).))))..)).......... ( -21.86) >DroYak_CAF1 22919 119 - 1 UGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCCACUUCCACAUCCACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUACUCCAACGCACAUUGUCGCAAAAUAUGAC .(((((((...)))))))((.(((.....))).......................................((((..(((.................))).))))..)).......... ( -18.33) >consensus UGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC____________ACUUUUCCCCAGCCACCCAAUCUUGCCCAGCUUAUUCUCCAACGCACAUUGUCGCAAAAUAUGAC .(((((((...)))))))((.(((.....))).......................................((((..(((.................))).))))..)).......... (-18.67 = -18.67 + -0.00)

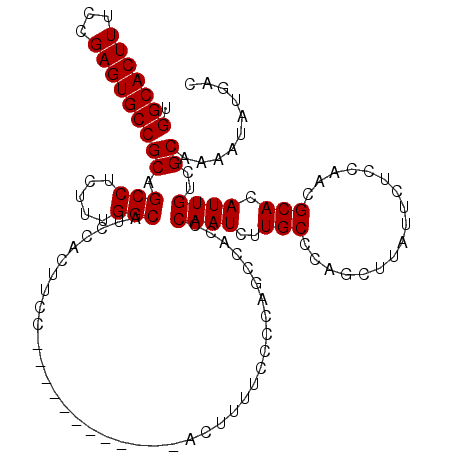
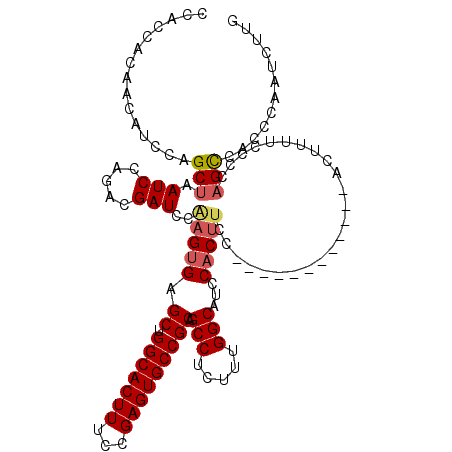
| Location | 12,899,701 – 12,899,808 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.24 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12899701 107 - 23771897 CCACCACAACAUCCAGCUAAUCCAGACGAUCCAAGUGAGCUGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC------------ACUUUUCCCCAGCCACCCAAUCUUG ...............(((.(((.....)))..(((((.((.(((((((...))))))))).(((.....)))...)))))..------------..........)))............ ( -22.20) >DroSec_CAF1 20681 107 - 1 CCACCACAACAUCCAGCUAAUCCAGACGAUCCAAGUGAGCUGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC------------ACUUUUCCCCAGCCACCCAAUCUUG ...............(((.(((.....)))..(((((.((.(((((((...))))))))).(((.....)))...)))))..------------..........)))............ ( -22.20) >DroSim_CAF1 18518 107 - 1 CCACCACAACAUCCAGCUAAUCCAGACGAUCCAAGUGAGCUGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC------------ACUUUUCCCCAGCCACCCAAUCUUG ...............(((.(((.....)))..(((((.((.(((((((...))))))))).(((.....)))...)))))..------------..........)))............ ( -22.20) >DroEre_CAF1 25941 107 - 1 CCACCACAACAGCCAGCUAAUCCAGACGAUCCAAGUGAGCUGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC------------ACUUUUCCCCAGCCACCCAAUCUUG ...............(((.(((.....)))..(((((.((.(((((((...))))))))).(((.....)))...)))))..------------..........)))............ ( -22.20) >DroYak_CAF1 22959 119 - 1 CCACCACAACAUCCAGCUAAUCCAGACGAUCCUAGUGAGCUGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCCACUUCCACAUCCACUUUUCCCCAGCCACCCAAUCUUG ...........................(((....(((.((((((((((...)))))))...(((.....)))................................))))))...)))... ( -21.70) >DroAna_CAF1 48022 107 - 1 CCACCACAACAUCCAGCUAAUCCAGACGAUCUGAGUGAGCUGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCCCAUCCCUAU--------ACCUUUCUCCUGUUUC----UUUUU .......((((...((((.(..(((.....)))..).))))(((((((...)))))))...(((.....)))..............--------..........))))..----..... ( -22.90) >consensus CCACCACAACAUCCAGCUAAUCCAGACGAUCCAAGUGAGCUGGCACUUUCCGAGUGCCGCAGCCUCUUUGGCAUCCACUUCC____________ACUUUUCCCCAGCCACCCAAUCUUG ...............(((.(((.....)))..(((((.((.(((((((...))))))))).(((.....)))...)))))........................)))............ (-19.48 = -19.90 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:51 2006