
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,899,480 – 12,899,595 |
| Length | 115 |
| Max. P | 0.914314 |

| Location | 12,899,480 – 12,899,595 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -32.69 |
| Consensus MFE | -19.04 |
| Energy contribution | -20.46 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12899480 115 + 23771897 AACCACUGUCAUAGUUUAUCACAUUCUGUGCAGUGGGAGGAAUUUUUUAUUCCUCCGCUUGCUUGGCUACUUGUGCCA-UUUUCUUCUUCCCACAAAA----AAAGAAAGUGUACCAGGG ...(((((.(((((...........))))))))))((((((((.....))))))))(((.....)))..((.((((.(-(((((((.((.......))----.)))))))))))).)).. ( -35.30) >DroPse_CAF1 28844 93 + 1 GAAGACUGUCAUAGUUUAUCACAUUCUGUGCAGUGGGAAGCAUUUUUUAUUUCUCC--------GGCUACUUGCAC-U-UUUUGUACCGCACACAG----------AAGCUGU------- ((((((((...)))))).))...(((((((..((((..(((...............--------.)))...((((.-.-...)))))))).)))))----------)).....------- ( -17.69) >DroSim_CAF1 18289 114 + 1 AACCACUGUCAUAGUUUAUCACAUUCUGUGCAGUGGGAGGAAUUUUUUAUUCCACCGCUUGCUUGGCUACUUGUGC-G-UCUUGUUCUUCCCACAAAA----AAAGAAAGUGUGCCAGGG ..((((((.(((((...........)))))))))))..(((((.....)))))........(((((((((((....-.-..((((.......))))..----.....))))).)))))). ( -33.24) >DroEre_CAF1 25735 109 + 1 GUACACUGUCAUAGUUUAUCACAUUCUGUGCAGUGGGAGGAAUUUUUUAUUCCUCCGC----UCGGCUACUUGUAC-G-UUUUGUUCUACCCACAAAA----AA-GAAAGUGUGCCGGGG ...(((((.(((((...........))))))))))((((((((.....)))))))).(----((((((((((....-.-((((((.......))))))----..-..))))).)))))). ( -41.60) >DroYak_CAF1 22748 111 + 1 GAACACUGUCAUAGUUUAUCACAUUCUGUGCAGUGGGAGGAAUUUUUUAUUCCUCCGC----UUGGCUACUUGUAC-GCUUUUGUUCUACCCACAAAA----AACGAAAGUGUGCCAGGG ...(((((.(((((...........))))))))))((((((((.....)))))))).(----(((((.......((-(((((((((............----))))))))))))))))). ( -41.01) >DroAna_CAF1 47830 114 + 1 GAACACUGUCAUAGUUUAUCACAUUCUGUGCAGUGGGUAGAAUUU-UUAUUUCUCCGCU---UUGGAUACUUGUGC-U-UUUUGUUCCUCCCACAUAGUGUGCAAAAAAGUGUCAUGGGG ...(((((.(((((...........))))))))))((.((((...-....)))))).((---...(((((((.(((-.-...(((.......)))......)))...)))))))..)).. ( -27.30) >consensus GAACACUGUCAUAGUUUAUCACAUUCUGUGCAGUGGGAGGAAUUUUUUAUUCCUCCGC____UUGGCUACUUGUAC_G_UUUUGUUCUUCCCACAAAA____AAAGAAAGUGUGCCAGGG ...(((((.(((((...........))))))))))((((((((.....))))))))...........(((((.......((((((.......)))))).........)))))........ (-19.04 = -20.46 + 1.42)
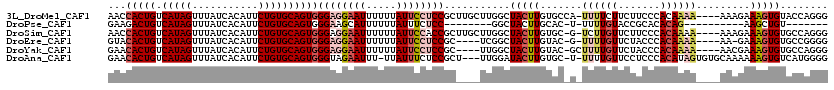
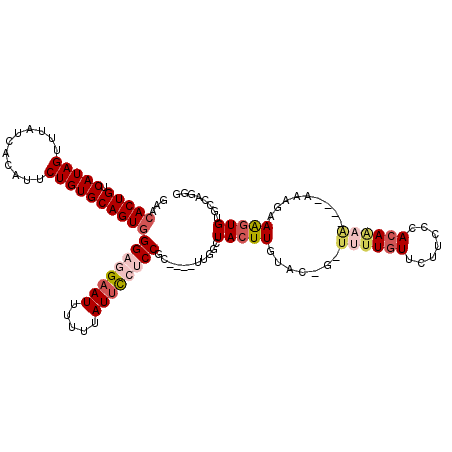

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:48 2006