| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,898,798 – 12,898,894 |
| Length | 96 |
| Max. P | 0.865075 |

| Location | 12,898,798 – 12,898,894 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -13.31 |
| Energy contribution | -12.82 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.865075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
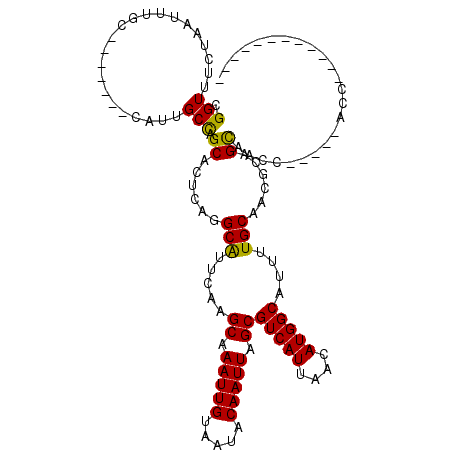
>3L_DroMel_CAF1 12898798 96 - 23771897 CGGUUUCUAAUUUGC-------CAUUGCCAGCACUCAGGCAUUCAAGCAAAUUGUAAUACAAUUAGCGUCAUUAACAUGGCAUUUUGCAACGCAGCAACC-----AGC------------ .((((.((...((((-------...(((((((......))......((.(((((.....))))).))..........)))))....))))...)).))))-----...------------ ( -23.40) >DroPse_CAF1 28238 116 - 1 CGGUUUCUAAUUUGCCAUUUGCCAUUGCUUGCAUUUACGCAUUCAAGCAAAUUGUAAUACAAUUAGCGUCAUUUACAUGGCAUUUUGCAUGCCAGCCAAC----UGCUUGCAUACACCCA .(((.........((.(..((((((((..(((......)))..)))((.(((((.....))))).))..........)))))..).))((((.(((....----.))).))))..))).. ( -28.60) >DroSec_CAF1 19726 97 - 1 CGGUUUCUAAUUUGC-------CAUUGCCAGCACUCAGGCAUUCAAGCAAAUUGUAAUACAAUUAGCGUCAUUAACAUGGCAUUUUGCAACGCAGCAACC----CAUC------------ .((((.((...((((-------...(((((((......))......((.(((((.....))))).))..........)))))....))))...)).))))----....------------ ( -22.50) >DroSim_CAF1 17567 96 - 1 CGGUUUCUAAUUUGC-------CAUUGCCAGCACUCAGGCAUUCAAGCAAAUUGUAAUACAAUUAGCGUCAUUAACAUGGCAUUUUGCAACGCAGCAACU-----AGC------------ ............(((-------(((((((........)))).....((.(((((.....))))).)).........))))))..((((......))))..-----...------------ ( -20.80) >DroAna_CAF1 47260 101 - 1 CGGUUCCUAAUUUUC-------CAUUGCCUGCUCUCGGGCGUUGAAGCAAAUUGUAAUACAAUUAGCGUCAUUUACAUGGCAUUUUGCAACGCAGUCAGGACUCCCUC------------ .((.((((.......-------....(((((....)))))((((..((.(((((.....))))).))(((((....)))))......))))......))))..))...------------ ( -25.90) >DroPer_CAF1 23273 116 - 1 CGGUUUCUAAUUUGCCAUUUGCCAUUGCUUGCAUUUACGCAUUCAAGCAAAUUGUAAUACAAUUAGCGUCAUUUACAUGGCAUUUUGCAUGCCAGCCAAC----UGCUUGCAUACACCCA .(((.........((.(..((((((((..(((......)))..)))((.(((((.....))))).))..........)))))..).))((((.(((....----.))).))))..))).. ( -28.60) >consensus CGGUUUCUAAUUUGC_______CAUUGCCAGCACUCAGGCAUUCAAGCAAAUUGUAAUACAAUUAGCGUCAUUAACAUGGCAUUUUGCAACGCAGCAACC_____ACC____________ .(((......................))).((......(((.....((.(((((.....))))).))(((((....)))))....)))......))........................ (-13.31 = -12.82 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:47 2006