
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,889,959 – 12,890,062 |
| Length | 103 |
| Max. P | 0.848575 |

| Location | 12,889,959 – 12,890,062 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -8.42 |
| Energy contribution | -8.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12889959 103 + 23771897 UGCAUUGUUUUCAAAAUGCUUCGAGUAACUUUUGCAGAAUGGCUGCUGAACAGCC-AAAGCCAAGGC--AAAA---GCAAAAGUUAAGCGCCCAACAAAAUGGAAUUUA .(((((........)))))..((..((((((((((....((((((.....)))))-)..((....))--....---))))))))))..)).(((......)))...... ( -33.30) >DroVir_CAF1 26767 80 + 1 UGCAUUGUUUGCAAAAUGCUUCAAGUAAAUUAUGCAGAAUUGC--------------------------AGAA---GCGAAAGUUAAGCGCACAACAAAGUGGAAUUUA .((....(((((((..(((..............)))...))))--------------------------)))(---((....)))..)).(((......)))....... ( -16.44) >DroSim_CAF1 14068 87 + 1 UGCAUUGUUUUCAAAAUGCUUCGAGUAACUUUUGCA----------------ACC-AAAGCCAAGGC--AAAA---GCAAAAGUUAAGCGCCCAACAAAAUGGAAUUUA .(((((........)))))..((..((((((((((.----------------...-...((....))--....---))))))))))..)).(((......)))...... ( -20.90) >DroEre_CAF1 21364 106 + 1 UGCAUUGUUUGCAAAAUGCUUCGAGUAACUUAUGCAGAAUGGCUGUUGAGCAGCC-AAAGCCAAGGC--AACAGCAGCAAAAGUUAAGCGCCCAACAAAAUGGAAUUUA .(((((........)))))..((..((((((.(((....((((((.....)))))-)..((....))--.......))).))))))..)).(((......)))...... ( -30.90) >DroYak_CAF1 18276 104 + 1 UGCAUUGUUUGCAAAAUGCUUCGAGUAACUUAUGCAGAAUGGCUGCUGAACAGCCGAAAGCCAAGGC--AACU---GCAAAAGUUAAGCGCCCAACAAAAUGGAAUUUA .(((((........)))))..((..((((((.(((((..((((((.....))))))...((....))--..))---))).))))))..)).(((......)))...... ( -33.00) >DroAna_CAF1 43822 94 + 1 UGCAUUGUUUGCAAAAUGCUCCGACUAAAUUAUGCAGAAUUGCUGCU------------GCCAAGGAGUCGAA---GCCAAAGUUAAGCGCCCAGCAAAAUGCAAUUUG ((((((..((((.....(((.(((((.......((((.....)))).------------.......))))).)---))....(.....).....))))))))))..... ( -23.56) >consensus UGCAUUGUUUGCAAAAUGCUUCGAGUAACUUAUGCAGAAUGGCUGCU_____GCC_AAAGCCAAGGC__AAAA___GCAAAAGUUAAGCGCCCAACAAAAUGGAAUUUA .(((((........)))))..((..((((((.(((.........................................))).))))))..))................... ( -8.42 = -8.98 + 0.56)

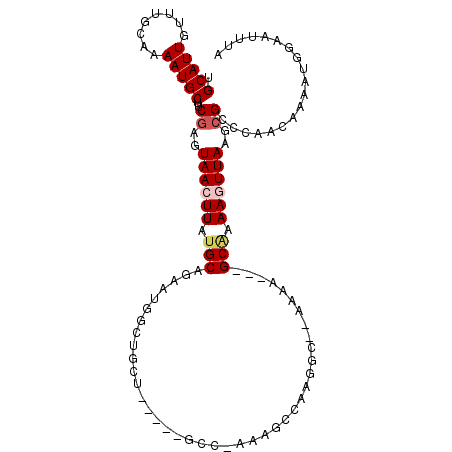
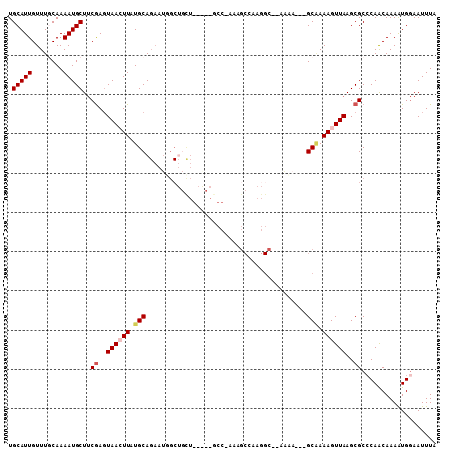
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:44 2006