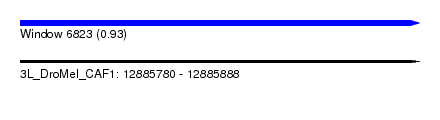
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,885,780 – 12,885,888 |
| Length | 108 |
| Max. P | 0.931725 |

| Location | 12,885,780 – 12,885,888 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.45 |
| Mean single sequence MFE | -30.11 |
| Consensus MFE | -12.46 |
| Energy contribution | -14.28 |
| Covariance contribution | 1.82 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12885780 108 + 23771897 GCAUUUCCUGAAAAUAAACACCUGUCAUUCAGGUGGGCGAAUGGAAGUGUGAGG--------AAGUGUAGCCUAUUUGCAAGAAAGGGG-GUACAAAUUAUUUCCAGCAG--CGGAAAG ...(((((..........((((((.....)))))).((...((((((((.....--------...((((.(((.(((.....))).)))-.))))...))))))))...)--)))))). ( -33.30) >DroVir_CAF1 24621 89 + 1 GAAUUU-CCGAAAAUAAACACCUGUCUUUAAU---------UG---------GA--------AAGCCCACCCAAUUCG--ACCGCC-CAGCUGCAAUUGAUUUACAGGGGGCUGUACAU ....((-((((......((....))......)---------))---------))--------)(((((.((....(((--(..((.-.....))..))))......)))))))...... ( -17.80) >DroSec_CAF1 11953 107 + 1 GCAUUUCCUGAAAAUAAACACCUGUCAUUCAGGUGGGCGAAUGGAAGUGUGAGG--------AAGUGCAGCCUAUUUGCAAGAACG-UU-GUACAAAUUAUUUCCAGCAG--CGGAAAG ...(((((..........((((((.....)))))).((...((((((((.....--------..(((((((..............)-))-))))....))))))))...)--)))))). ( -31.64) >DroSim_CAF1 9746 107 + 1 GCAUUUCCUGAAAAUAAACACCUGUCAUUCAGGUGGGCGAAUGGCAGUGUGAGG--------AAGUGCAGCCUAUUAGCAAGAACG-UU-GUACAAAUUAUUUCCACAAG--CGGAAAG (((((((((.........((((((((((((........))))))))).))))))--------)))))).((......)).......-..-..........(((((.....--.))))). ( -32.10) >DroEre_CAF1 16863 115 + 1 GCAUUUUCCGAAAAUAAACACCUGUCAUUCAGGUGGGCGAAUGGGAGUGUGGGAAGUGUGUGAAGUGCAGCCUCUUCGCAAGAAUG-UA-GUACAAAUUAUUUCCAGCAG--CGGAAAG ....((((((........(((((.((((((........)))))).)).)))(((((((......((((.((...(((....))).)-).-))))....))))))).....--)))))). ( -33.50) >DroYak_CAF1 13136 95 + 1 GCAUUUUCGGAAAAUAAACACCUGUCAUUCAGGUGGGCGAUGGGGAGUGGGAGAACUG--------------------CGAGAACG-CA-GUACAAAUUAUUUCCAGCAG--CGGAAAG ...((((((.........((((((.....)))))).((....((((((((..(.((((--------------------((....))-))-)).)...)))))))).))..--)))))). ( -32.30) >consensus GCAUUUCCUGAAAAUAAACACCUGUCAUUCAGGUGGGCGAAUGGAAGUGUGAGA________AAGUGCAGCCUAUUCGCAAGAACG_UA_GUACAAAUUAUUUCCAGCAG__CGGAAAG ((((((((..........((((((.....)))))).......))))))))..................................................(((((........))))). (-12.46 = -14.28 + 1.82)

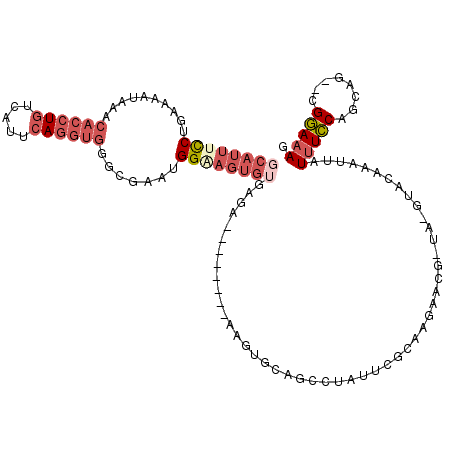
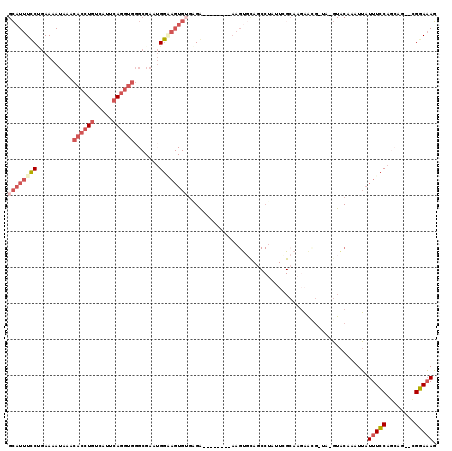
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:43 2006