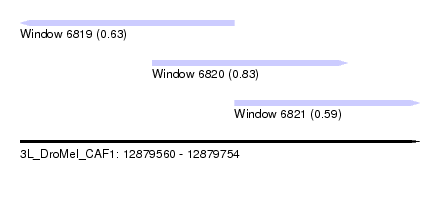
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,879,560 – 12,879,754 |
| Length | 194 |
| Max. P | 0.832845 |

| Location | 12,879,560 – 12,879,664 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -23.51 |
| Energy contribution | -23.27 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12879560 104 - 23771897 UACAGCAAUGGAUGUGGAGUAUCCUGUGGAUACAUGUGUAUGGGAUGGCAUUGGAUAAUAGUGUCUUGCAGUCAAAGUGCGUGUAUGCAAGGAAUUAUCAAGUG ((((........))))((...((((.((.(((((((..(....(((.(((..(((((....)))))))).)))...)..))))))).))))))....))..... ( -26.90) >DroSec_CAF1 6487 104 - 1 UACAGCAAUGGAAGUGGAGUAUCCUGUGGAUACAUGUGUAUGGGAUGGCAUUGGAUAAUAGUGUCUUGCAGUCAAAGUGCGUGUAUGCAAGGAAUUAUCAAGUG ..((....))......((...((((.((.(((((((..(....(((.(((..(((((....)))))))).)))...)..))))))).))))))....))..... ( -26.70) >DroSim_CAF1 4046 104 - 1 UACAGCAAUGGAAGUGGAGUAUCCUGUGGAUACAUGUGUAUGGGAUGGCAUUGGAUAAUAGUGUCUUGCAGUCAAAGUGCGUGUAUGCAAGGAAUUAUCAAGUG ..((....))......((...((((.((.(((((((..(....(((.(((..(((((....)))))))).)))...)..))))))).))))))....))..... ( -26.70) >DroEre_CAF1 9254 104 - 1 CACAGCAAUGGAAGUGGCGUAUCCUGUGGAUACAUGUGUAUGGGAUGGCAUUGGAUAAUAGUGUCUUGCAGUCAAAGUGCGUGUAUGCAAGGAAUUAUCAAGUG (((.((.((....)).))...((((.((.(((((((..(....(((.(((..(((((....)))))))).)))...)..))))))).))))))........))) ( -27.70) >DroYak_CAF1 5924 104 - 1 CACGGCAAUGGAAGUGGAGUAUCCUGUGGAUACAUGUGUACGGGAUGGCAUUGGAUAAUAGUGUCUUGCAGUCAAAGUGCGUGUAUGCAAGGAAUUAUCAAGUG (((..(....)..)))((...((((.((.(((((((..(....(((.(((..(((((....)))))))).)))...)..))))))).))))))....))..... ( -27.70) >DroAna_CAF1 31301 102 - 1 CAGAGCA--GGAGGUCGAGUAUCUGGCGGAUACAUGCGAAUACGAUGGCAAUGGAUAAUAGUGUCUUGCAGUCAAAGUGCGUGUAUGCAGGGAAUUACCAAGUG ....((.--(((.........))).))(.(((((((((.....(((.(((..(((((....)))))))).)))....))))))))).).((......))..... ( -26.40) >consensus CACAGCAAUGGAAGUGGAGUAUCCUGUGGAUACAUGUGUAUGGGAUGGCAUUGGAUAAUAGUGUCUUGCAGUCAAAGUGCGUGUAUGCAAGGAAUUAUCAAGUG ....(((...........((((((...))))))(..(((((..(((.(((..(((((....)))))))).)))...)))))..).)))................ (-23.51 = -23.27 + -0.25)

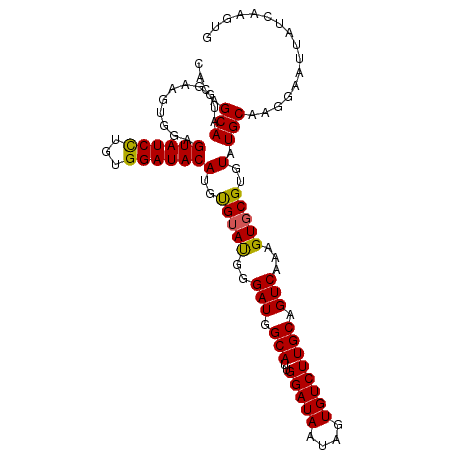
| Location | 12,879,624 – 12,879,719 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -20.30 |
| Energy contribution | -22.30 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12879624 95 + 23771897 UACACAUGUAUCCACAGGAUACUCCACAUCCAUUGCUGUACGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUUCUCGUCUGCGGCUUUGAAUUAU .......((((((...))))))........((..((((((((..((((((((((....)))))).))))......)))..)))))..))...... ( -30.70) >DroSec_CAF1 6551 95 + 1 UACACAUGUAUCCACAGGAUACUCCACUUCCAUUGCUGUACGUACUCUGAUGCCCCUGGGCAUAUGGAGCCUUCUCGUCUGCGGCUUUGAAUUAU .......((((((...))))))........((..((((((((..((((((((((....))))).)))))......)))..)))))..))...... ( -26.80) >DroSim_CAF1 4110 95 + 1 UACACAUGUAUCCACAGGAUACUCCACUUCCAUUGCUGUACGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUUCCCGUCUGCGGCUUUGAAUUAU .......((((((...))))))........((..((((((((..((((((((((....)))))).))))......)))..)))))..))...... ( -30.00) >DroEre_CAF1 9318 95 + 1 UACACAUGUAUCCACAGGAUACGCCACUUCCAUUGCUGUGCGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUGCUCGUCUGCGGCUUUGAAUUAU ..........((((.((..((((((((.......).)).)))))..))((((((....))))))))))((((((......))))))......... ( -32.50) >DroYak_CAF1 5988 95 + 1 UACACAUGUAUCCACAGGAUACUCCACUUCCAUUGCCGUGAGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUGUUCGGCUGCGGCUUUGAAUUAU .......((((((...))))))........((..(((((.....((((((((((....)))))).))))(((....))).)))))..))...... ( -34.50) >DroAna_CAF1 31365 73 + 1 UUCGCAUGUAUCCGCCAGAUACUCGACCUCC--UGCUCUG------CUCAUCCUCCUGA------------UUUUAUUC--UGGCUUUGAAUUAU .............((((((.....((.(...--.).))..------.(((......)))------------......))--)))).......... ( -9.30) >consensus UACACAUGUAUCCACAGGAUACUCCACUUCCAUUGCUGUACGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUUCUCGUCUGCGGCUUUGAAUUAU .......(((((.....)))))........((..((((((((..((((((((((....)))))).))))......)))..)))))..))...... (-20.30 = -22.30 + 2.00)



| Location | 12,879,664 – 12,879,754 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.75 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12879664 90 + 23771897 CGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUUCUCGUCUGCGGCUUUGAAUUAUUUAAAAUUGUUUUCUCACGC---C-CGCUUUUGCGGGUG ....((((((((((....)))))).))))(((.(......).)))...........................(((---(-(((....))))))) ( -31.00) >DroSec_CAF1 6591 90 + 1 CGUACUCUGAUGCCCCUGGGCAUAUGGAGCCUUCUCGUCUGCGGCUUUGAAUUAUUUAAAAUUGUUUUCUCACGC---C-CGCUCUUGCGGGUG .........(((((....)))))..((((((...........))))))........................(((---(-(((....))))))) ( -26.80) >DroSim_CAF1 4150 90 + 1 CGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUUCCCGUCUGCGGCUUUGAAUUAUUUAAAAUUGUUUUCUCACGC---C-CGCUCCUGCGGGUG ....((((((((((....)))))).)))).....(((....)))............................(((---(-(((....))))))) ( -31.80) >DroEre_CAF1 9358 90 + 1 CGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUGCUCGUCUGCGGCUUUGAAUUAUUUAAAAUUGUUUCCUCAUAC---C-CGCUCUUGUGGGUG ....((((((((((....)))))).))))(((((......)))))...........................(((---(-(((....))))))) ( -33.60) >DroYak_CAF1 6028 93 + 1 AGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUGUUCGGCUGCGGCUUUGAAUUAUUUAAAACUGUUUCGCCACACCCCC-CGCUCUUCUGGGUG (((.((((((((((....)))))).)))))))....(((.((((.(((((.....))))).))))...))).(((((..-.........))))) ( -31.60) >DroAna_CAF1 31403 71 + 1 ------CUCAUCCUCCUGA------------UUUUAUUC--UGGCUUUGAAUUAUUUAAAAUUGUUUCUUGCCGC---CACACUUUGUUGGGGG ------......((((..(------------(.......--((((...(((.((........)).)))..)))).---........))..)))) ( -12.69) >consensus CGUACUCUGAUGCCCCUGGGCAUCUGGAGGCUUCUCGUCUGCGGCUUUGAAUUAUUUAAAAUUGUUUCCUCACGC___C_CGCUCUUGCGGGUG ....((((((((((....)))))).))))....(((((..(((..(((((.....)))))..(((......)))......)))....))))).. (-13.16 = -13.75 + 0.59)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:41 2006