
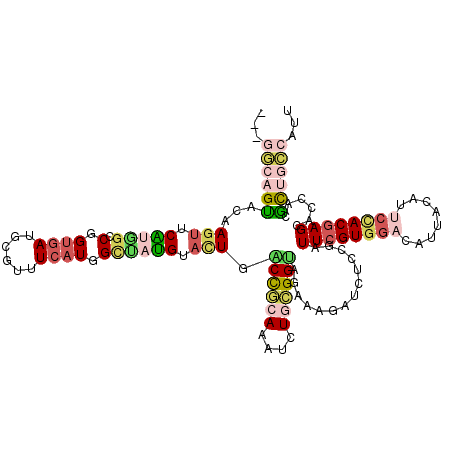
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,877,161 – 12,877,277 |
| Length | 116 |
| Max. P | 0.975837 |

| Location | 12,877,161 – 12,877,277 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -25.38 |
| Energy contribution | -26.88 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
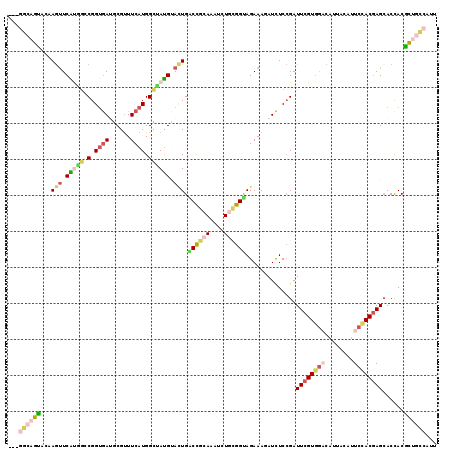
>3L_DroMel_CAF1 12877161 116 + 23771897 ---GGCAGUAAAAGUUCGUGGCCGGUGAUGCGUUUCAUGGCUAUGUACUGGCCGCAAAUCUGCGGUAGAAAGAUCUCCGAUUCGUGGACAUUACAUUCUACGAGCACUACGCUGCCAUU ---((((((........((((((((((..((........))....)))))))))).....(((((.(((....)))))...(((((((........))))))))))....))))))... ( -44.70) >DroVir_CAF1 13279 119 + 1 ACACAUUGCGCCAACUCGCCAGCGGAGACGCGCUUCAUGGCAACGCAAUGACUAGAAAUUUGAGGCAGAAAUAUCUCAGAUUCGUAGCCAUUACAUUCCACAAGCACCACGUUUCCGUU .......(((......))).((((((((((.((((..(((.((.(.((((.(((..((((((((..(....)..))))))))..))).)))).).))))).))))....)))))))))) ( -35.40) >DroSec_CAF1 4150 116 + 1 ---GGCAGUAAAAGUUCGCGGCCGGUGAUGCGUUUCAUGGCUAUGUACUGACCGCAAAUCUGCGGUAGAAAGAUCUCCGAUUCGUGGACAUUGCAUUCCACGAGCACUACGCUGCCAUU ---((((((...(((.((..((((.(((......)))))))..)).))).((((((....))))))............(.((((((((........))))))))).....))))))... ( -42.10) >DroEre_CAF1 5135 116 + 1 ---GGCAGUACAAGUUCAUGGGCGGUGAUGCGUUUCAUGGUUAUGUACUGACCGCAAACCUGCGGUAGAAAUAUCUCCGAUUCGUGGACAUUACAUUCCACGAGCACCACGCUGCCAUU ---((((((....(((....))).(((.(((......(((..((((....((((((....))))))....))))..)))..(((((((........)))))))))).)))))))))... ( -43.30) >DroYak_CAF1 1714 116 + 1 ---GGCAGUACAAGUUCAUGGCCGGUGAUGCGUUUCAUGGCCAUGUACUGGCCGCAAAUUUGCGGUAGAAAGAUCUCCGAUUCGUGGACAUUACAUUCCACGAGCACCACGCUGCCAUU ---((((((...(((.((((((((.(((......))))))))))).))).((((((....))))))............(.((((((((........))))))))).....))))))... ( -48.50) >DroAna_CAF1 2429 116 + 1 ---CACUGGAUUAGUUCAUUGGCGGUAACGCGUUUCAUGGCAAUGAAUUGGCCAGAAAGAUAUGGCAGAAAUAUUUCCGAUUCGUGACCACUAGACUCCACGAGGACCACUUUUUCGUU ---....(((...........(((....)))((((..(((..((((((((((((........)))).(((....)))))))))))..)))..)))))))(((((((.....))))))). ( -31.10) >consensus ___GGCAGUACAAGUUCAUGGCCGGUGAUGCGUUUCAUGGCUAUGUACUGACCGCAAAUCUGCGGUAGAAAGAUCUCCGAUUCGUGGACAUUACAUUCCACGAGCACCACGCUGCCAUU ...((((((...(((.(((((.(.((((......)))).)))))).))).((((((....))))))..............((((((((........))))))))......))))))... (-25.38 = -26.88 + 1.51)

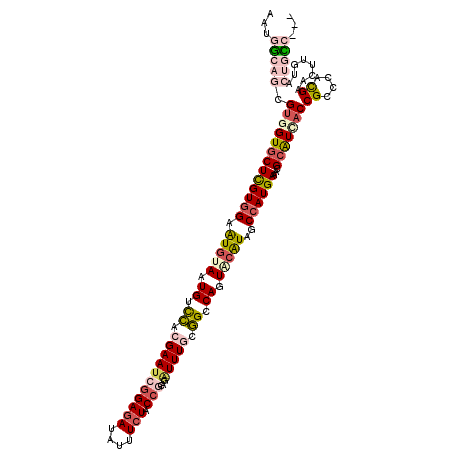
| Location | 12,877,161 – 12,877,277 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -37.47 |
| Consensus MFE | -22.39 |
| Energy contribution | -24.28 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
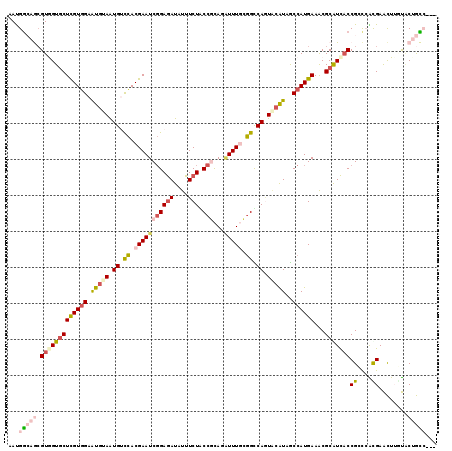
>3L_DroMel_CAF1 12877161 116 - 23771897 AAUGGCAGCGUAGUGCUCGUAGAAUGUAAUGUCCACGAAUCGGAGAUCUUUCUACCGCAGAUUUGCGGCCAGUACAUAGCCAUGAAACGCAUCACCGGCCACGAACUUUUACUGCC--- ...(((((...(((..((((...(((((.((.((.(((((((((((....))).))...)))))).)).)).))))).(((.(((......)))..))).)))))))....)))))--- ( -32.90) >DroVir_CAF1 13279 119 - 1 AACGGAAACGUGGUGCUUGUGGAAUGUAAUGGCUACGAAUCUGAGAUAUUUCUGCCUCAAAUUUCUAGUCAUUGCGUUGCCAUGAAGCGCGUCUCCGCUGGCGAGUUGGCGCAAUGUGU .(((....))).(((((((((((((((((((((((.((((.((((..........)))).)))).))))))))))))).)))).)))))).....(((..(((......)))...))). ( -45.70) >DroSec_CAF1 4150 116 - 1 AAUGGCAGCGUAGUGCUCGUGGAAUGCAAUGUCCACGAAUCGGAGAUCUUUCUACCGCAGAUUUGCGGUCAGUACAUAGCCAUGAAACGCAUCACCGGCCGCGAACUUUUACUGCC--- ...(((((.....((((((((((........)))))))...(((((....))).)))))..(((((((((.((.....((........))...)).)))))))))......)))))--- ( -37.40) >DroEre_CAF1 5135 116 - 1 AAUGGCAGCGUGGUGCUCGUGGAAUGUAAUGUCCACGAAUCGGAGAUAUUUCUACCGCAGGUUUGCGGUCAGUACAUAACCAUGAAACGCAUCACCGCCCAUGAACUUGUACUGCC--- ...(((((.(((((((((((((.(((((.((.((.(((((((((((....))).)))...))))).)).)).)))))..))))))...)))))))....((......))..)))))--- ( -40.50) >DroYak_CAF1 1714 116 - 1 AAUGGCAGCGUGGUGCUCGUGGAAUGUAAUGUCCACGAAUCGGAGAUCUUUCUACCGCAAAUUUGCGGCCAGUACAUGGCCAUGAAACGCAUCACCGGCCAUGAACUUGUACUGCC--- .(((((.(.((((((((((((((........))))))).(((((((....))).))(((....)))(((((.....)))))..))...)))))))).)))))..............--- ( -44.40) >DroAna_CAF1 2429 116 - 1 AACGAAAAAGUGGUCCUCGUGGAGUCUAGUGGUCACGAAUCGGAAAUAUUUCUGCCAUAUCUUUCUGGCCAAUUCAUUGCCAUGAAACGCGUUACCGCCAAUGAACUAAUCCAGUG--- .........(((((...((((...((..(((((...((((.((((....))))((((........))))..))))...)))))))..))))..)))))..................--- ( -23.90) >consensus AAUGGCAGCGUGGUGCUCGUGGAAUGUAAUGUCCACGAAUCGGAGAUAUUUCUACCGCAGAUUUGCGGCCAGUACAUAGCCAUGAAACGCAUCACCGCCCACGAACUUGUACUGCC___ ...(((((.(((((((((((((.(((((.((.((.(((((((((((....))).)))...))))).)).)).)))))..))))))...)))))))((....))........)))))... (-22.39 = -24.28 + 1.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:38 2006