
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,865,228 – 12,865,486 |
| Length | 258 |
| Max. P | 0.901531 |

| Location | 12,865,228 – 12,865,348 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -21.88 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.672599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12865228 120 + 23771897 CCACCUCAGCCCAACCCACAGUACGCAAUUCAAUACCAGGCCAACCGGCAAUAUGCCAUGCAAGGACAACCCAAUUCGGCCAAUCCUUGGCUCCACUUCUCAGUAGGGAUGUCCUAUCCA ........................(((...........((....))(((.....))).))).((((((.(((.....(((((.....)))))..(((....))).))).))))))..... ( -29.30) >DroSec_CAF1 103669 120 + 1 CCACCUCAGCCCAAUCCACAGUACGCAAUUCAAUACCAGGCCAACCGGCAAUAUGCCAUGCAAGGAAAACCCAACUCGGCCAAUCCUGGGCUCUACUUCUCAGUGGGGAUGUCCUAUCCA .......((((((.......(....)............((((....(((.....)))......((.....)).....)))).....))))))(((((....)))))(((((...))))). ( -30.30) >DroEre_CAF1 109017 120 + 1 CCACCUCAGCCCAACCCGCAAUACGCGAUUCAAUACCAGGCCAACAGGCAGUUCGCCCUGCAAGGACAGCCCAACUGGGGCCAUCCUGGGCUCUACUUCUCCGUAGGGAUGUCCUAUCCA .......(((((.....(((....((((...........(((....)))...))))..))).((((..((((.....))))..)))))))))((((......))))(((((...))))). ( -37.64) >DroYak_CAF1 115036 120 + 1 CCACCUCAGCUCAACCCACAGUACGCAAUUCAAUACCAGGCCCACAGGCAAUACGCCAUGCAAGGAGAUCCCAACUGGGCCAAUCCUGGGCUCUACUUCUCAAUGGGGAUGUCCUAUCCC ...(((((((...((.....))..))...........((((((...(((.....))).....((((...(((....)))....)))))))).)).........)))))............ ( -29.00) >consensus CCACCUCAGCCCAACCCACAGUACGCAAUUCAAUACCAGGCCAACAGGCAAUACGCCAUGCAAGGACAACCCAACUCGGCCAAUCCUGGGCUCUACUUCUCAGUAGGGAUGUCCUAUCCA .......(((((............(((...........(.....).(((.....))).))).((((...((......))....)))))))))((((......))))(((((...))))). (-21.88 = -22.12 + 0.25)


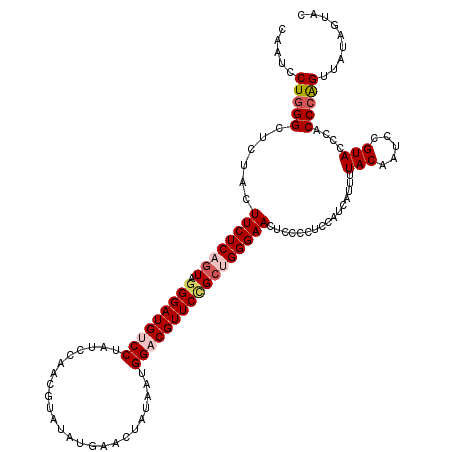
| Location | 12,865,268 – 12,865,388 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -25.81 |
| Energy contribution | -27.18 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12865268 120 + 23771897 CCAACCGGCAAUAUGCCAUGCAAGGACAACCCAAUUCGGCCAAUCCUUGGCUCCACUUCUCAGUAGGGAUGUCCUAUCCAACGUAUAUGAACUAUAAUGGACGUUCCGCUGGGAACUCAC ......(((.....)))...((((((....((.....))....)))))).......((((((((.(((((((((........................)))))))))))))))))..... ( -36.06) >DroSec_CAF1 103709 120 + 1 CCAACCGGCAAUAUGCCAUGCAAGGAAAACCCAACUCGGCCAAUCCUGGGCUCUACUUCUCAGUGGGGAUGUCCUAUCCAACGUAUAUGAGCUAUAAUGGACGUUCCGCUGGGAACUCCC ......(((.....)))..((.((((....((.....))....))))..)).....(((((((((((.((((((........................)))))))))))))))))..... ( -34.96) >DroEre_CAF1 109057 120 + 1 CCAACAGGCAGUUCGCCCUGCAAGGACAGCCCAACUGGGGCCAUCCUGGGCUCUACUUCUCCGUAGGGAUGUCCUAUCCAACGUACAUGAAAUGUUAUGGCCGUUCAGCUGGGAACUACC ......((.(((((.((..((.((((((.(((.(((((((((......))))))).......)).))).))))))....((((..(((((....)))))..))))..)).))))))).)) ( -42.11) >DroYak_CAF1 115076 120 + 1 CCCACAGGCAAUACGCCAUGCAAGGAGAUCCCAACUGGGCCAAUCCUGGGCUCUACUUCUCAAUGGGGAUGUCCUAUCCCACGUAUAUGAACUACAAUGGCCGUUCUGCCGGGAACUCCC ......((((..(((((((...((((.(((((....(((((.......)))))..(........)))))).)))).......(((.......))).)))).)))..))))(((....))) ( -39.90) >consensus CCAACAGGCAAUACGCCAUGCAAGGACAACCCAACUCGGCCAAUCCUGGGCUCUACUUCUCAGUAGGGAUGUCCUAUCCAACGUAUAUGAACUAUAAUGGACGUUCCGCUGGGAACUCCC ......(((.....)))..((.((((...((......))....))))..)).....((((((((.(((((((((........................)))))))))))))))))..... (-25.81 = -27.18 + 1.38)



| Location | 12,865,308 – 12,865,428 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -20.35 |
| Energy contribution | -21.79 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12865308 120 + 23771897 CAAUCCUUGGCUCCACUUCUCAGUAGGGAUGUCCUAUCCAACGUAUAUGAACUAUAAUGGACGUUCCGCUGGGAACUCACUUCCAUCAUUUUACAAUCCGUACCCACCCAGUUAUAGUAC ......((((......((((((((.(((((((((........................)))))))))))))))))................(((.....))).....))))......... ( -24.56) >DroSec_CAF1 103749 120 + 1 CAAUCCUGGGCUCUACUUCUCAGUGGGGAUGUCCUAUCCAACGUAUAUGAGCUAUAAUGGACGUUCCGCUGGGAACUCCCCUCCAUCAUUUUACAAUCCGUACCCACCCAGUUAUAGUAC .....(((((......(((((((((((.((((((........................)))))))))))))))))................(((.....)))....)))))......... ( -30.26) >DroEre_CAF1 109097 120 + 1 CCAUCCUGGGCUCUACUUCUCCGUAGGGAUGUCCUAUCCAACGUACAUGAAAUGUUAUGGCCGUUCAGCUGGGAACUACCCUUCAUCAUUUUACAAUCCGUAUCUACCCGGUUAUUGCAG .....(((((((((((......))))))...(((((.(.((((..(((((....)))))..))))..).)))))................................)))))......... ( -25.50) >DroYak_CAF1 115116 120 + 1 CAAUCCUGGGCUCUACUUCUCAAUGGGGAUGUCCUAUCCCACGUAUAUGAACUACAAUGGCCGUUCUGCCGGGAACUCCCCUCCAUCAUUUUACAAUCCGUAUCUGCCCAGUUAUAGUAC .....((((((..(((.........((((((...))))))..(((.((((........(((......)))(((....))).....))))..))).....)))...))))))......... ( -32.20) >consensus CAAUCCUGGGCUCUACUUCUCAGUAGGGAUGUCCUAUCCAACGUAUAUGAACUAUAAUGGACGUUCCGCUGGGAACUCCCCUCCAUCAUUUUACAAUCCGUACCCACCCAGUUAUAGUAC .....(((((......((((((((.(((((((((........................)))))))))))))))))................(((.....)))....)))))......... (-20.35 = -21.79 + 1.44)


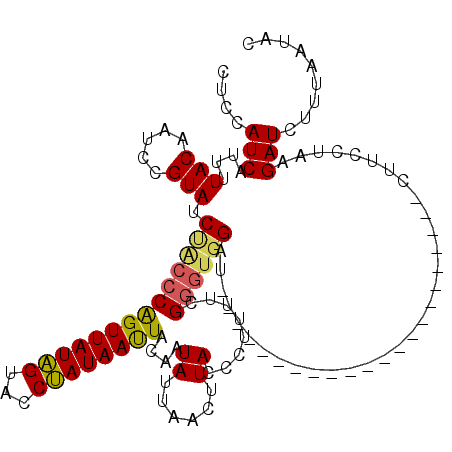
| Location | 12,865,388 – 12,865,486 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -15.80 |
| Consensus MFE | -8.24 |
| Energy contribution | -8.47 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12865388 98 + 23771897 UUCCAUCAUUUUACAAUCCGUACCCACCCAGUUAUAGUACCUAUAAUUACAAUAUUAACUUACCCUUUUCGGGUGGAU----------------------CUUCCUAAGAUCUUCGAUAC ....(((.................(((((((((((((...))))))))....((......))........)))))(((----------------------((.....)))))...))).. ( -16.00) >DroEre_CAF1 109177 120 + 1 CUUCAUCAUUUUACAAUCCGUAUCUACCCGGUUAUUGCAGCAAUAAUUACAAUAUUAACUUACCCUUUUAGUACUGCUAUAGUUAUGCAGUUUUAUUCUGCUUCCUAAGAUCUUUAAUAC ....(((..........(((........))).....((((.(((((...........(((.........)))(((((.........))))).))))))))).......)))......... ( -14.20) >DroYak_CAF1 115196 98 + 1 CUCCAUCAUUUUACAAUCCGUAUCUGCCCAGUUAUAGUACCUAUAACUACAAUAUUAACUUACCCUUUUCGGGUGGAU----------------------CUCCAUAGGAUCUUUAAUAC .(((.......(((.....)))((..(((((((((((...))))))))....((......))........)))..)).----------------------.......))).......... ( -17.20) >consensus CUCCAUCAUUUUACAAUCCGUAUCUACCCAGUUAUAGUACCUAUAAUUACAAUAUUAACUUACCCUUUUCGGGUGGAU______________________CUUCCUAAGAUCUUUAAUAC ....(((....(((.....))).((((((((((((((...))))))))....((......))........))))))................................)))......... ( -8.24 = -8.47 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:31 2006