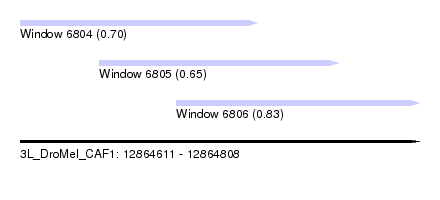
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,864,611 – 12,864,808 |
| Length | 197 |
| Max. P | 0.829413 |

| Location | 12,864,611 – 12,864,728 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -21.32 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12864611 117 + 23771897 CUGAGCGAUUUCAGUAACUCCGA-UACAUUUGCCACCCAUGCAGGCCAACGGCAGGCUAAUG--UUUUGGUUAUUGUUCAAGAUGGUUCCCAGCAACCUCAACAGCCAAAUCCUUGGCUA .((((((((..........((((-.(((((.(((...(.....)(((...))).))).))))--).))))..)))))))).((.((((......))))))...((((((....)))))). ( -31.60) >DroSec_CAF1 103051 120 + 1 CUUAACCAUUUCACUAACCCCCUUUCCUUUUGCCACCCCAGUCUGGCUUCGCCCCGCCAUGGGUUUUGGGUUUUUGUUCAAGAUGGUUCCCAGCAAUCUCAGCAGCCAAAUCCUUGGCUG ...((((((((((..(((((...........((((........))))...((((......))))...)))))..))....))))))))..............(((((((....))))))) ( -31.60) >DroEre_CAF1 108421 117 + 1 CUGACCGGUUUCGCUAACACCGA-UAUAUUGGCCACGCAUGCAGGGCAACGGCAGGCCAAUG--UUCUGGUUAUUGUACAAGAUGGUUCCCAACAAUCUCAACAGCCAAAUCCCUGGCUG ..(((((.....((....((((.-.(((((((((.....(((..(....).)))))))))))--)..))))....))......)))))..............((((((......)))))) ( -34.50) >DroYak_CAF1 114416 117 + 1 CUGACCGAUUUCAGUAACUCCGA-UGCAUUGGCCACGCAUGCAGGGCAACGGCAGGCCAAUG--UUCUGGUUAUUGUUCAAGAUGGUUCCCAACAACCUCAACAGCCAAAUCCCUGGCUG ......((...((((((((..((-.(((((((((.....(((..(....).)))))))))))--))).)))))))).))..((.((((......))))))..((((((......)))))) ( -39.70) >consensus CUGACCGAUUUCACUAACUCCGA_UACAUUGGCCACCCAUGCAGGGCAACGGCAGGCCAAUG__UUCUGGUUAUUGUUCAAGAUGGUUCCCAACAACCUCAACAGCCAAAUCCCUGGCUG ...........((((((((.......((((((((.....((.....))......))))))))......)))))))).....((.((((......))))))..((((((......)))))) (-21.32 = -22.07 + 0.75)



| Location | 12,864,650 – 12,864,768 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -25.92 |
| Energy contribution | -26.68 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
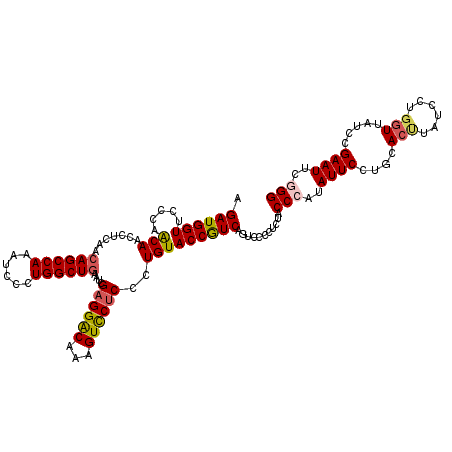
>3L_DroMel_CAF1 12864650 118 + 23771897 GCAGGCCAACGGCAGGCUAAUG--UUUUGGUUAUUGUUCAAGAUGGUUCCCAGCAACCUCAACAGCCAAAUCCUUGGCUAAAUCGAGGACAAAGUCCUCCCUGUACCAUCAGUCCGCUCU ((.((((((.((((......))--)))))))).........((((((...(((..........((((((....)))))).....((((((...)))))).))).)))))).....))... ( -35.00) >DroSec_CAF1 103091 120 + 1 GUCUGGCUUCGCCCCGCCAUGGGUUUUGGGUUUUUGUUCAAGAUGGUUCCCAGCAAUCUCAGCAGCCAAAUCCUUGGCUGAAUCGAGGGCAAAGUCCCCCCUGUACCGUCAGUCCCCUCU (.(((((...((((......))))...(((.((((((((.((((.((.....)).))))...(((((((....)))))))......)))))))).))).........))))).)...... ( -35.60) >DroEre_CAF1 108460 118 + 1 GCAGGGCAACGGCAGGCCAAUG--UUCUGGUUAUUGUACAAGAUGGUUCCCAACAAUCUCAACAGCCAAAUCCCUGGCUGAAUCGAGGACAAAGUCCUCCCUGUACCGUCAGUCCCCUCU ...((((.((((((((....((--(..(((..((..(.....)..))..)))))).......((((((......))))))....((((((...))))))))))..))))..))))..... ( -36.60) >DroYak_CAF1 114455 118 + 1 GCAGGGCAACGGCAGGCCAAUG--UUCUGGUUAUUGUUCAAGAUGGUUCCCAACAACCUCAACAGCCAAAUCCCUGGCUGAAUCGAGGACAAAGUUCUCCCUGUACCGUCAGUGCCCUCU ..(((((((((((((((((...--...)))...((((((..(((((((......))))....((((((......)))))).)))..)))))).......))))..))))...)))))).. ( -39.60) >consensus GCAGGGCAACGGCAGGCCAAUG__UUCUGGUUAUUGUUCAAGAUGGUUCCCAACAACCUCAACAGCCAAAUCCCUGGCUGAAUCGAGGACAAAGUCCUCCCUGUACCGUCAGUCCCCUCU ...((((.......(((((........))))).........((((((.....(((.......((((((......))))))....((((((...))))))..))))))))).))))..... (-25.92 = -26.68 + 0.75)

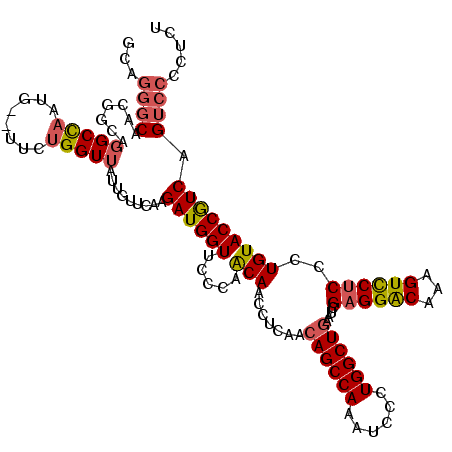
| Location | 12,864,688 – 12,864,808 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -24.14 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12864688 120 + 23771897 AGAUGGUUCCCAGCAACCUCAACAGCCAAAUCCUUGGCUAAAUCGAGGACAAAGUCCUCCCUGUACCAUCAGUCCGCUCUACCCAUAUUCCUGUACUUAUCCCCGUUAUCCGAAUUCGGG .((((((...(((..........((((((....)))))).....((((((...)))))).))).))))))...........(((..((((....((........)).....))))..))) ( -30.70) >DroSec_CAF1 103131 120 + 1 AGAUGGUUCCCAGCAAUCUCAGCAGCCAAAUCCUUGGCUGAAUCGAGGGCAAAGUCCCCCCUGUACCGUCAGUCCCCUCUACCCAUAUUCCUGCACUUAUCCUGGUUAUCCGAAUUCGGG .((((((...(((...((....(((((((....)))))))....))(((......)))..))).))))))...........(((..((((....(((......))).....))))..))) ( -29.60) >DroEre_CAF1 108498 120 + 1 AGAUGGUUCCCAACAAUCUCAACAGCCAAAUCCCUGGCUGAAUCGAGGACAAAGUCCUCCCUGUACCGUCAGUCCCCUCUACCCAUAUUCCUGCACCUAUCCUGGUUUCCCGAAUUCUGG .((((((...((..........((((((......))))))....((((((...))))))..)).))))))............(((.((((....(((......))).....))))..))) ( -30.70) >DroYak_CAF1 114493 120 + 1 AGAUGGUUCCCAACAACCUCAACAGCCAAAUCCCUGGCUGAAUCGAGGACAAAGUUCUCCCUGUACCGUCAGUGCCCUCUACCCAUAUUCCUGCAAUUAUCCUGGUUAUCCGAAUUCCGG .((((((...((..........((((((......))))))....((((((...))))))..)).))))))...............................((((...........)))) ( -24.40) >consensus AGAUGGUUCCCAACAACCUCAACAGCCAAAUCCCUGGCUGAAUCGAGGACAAAGUCCUCCCUGUACCGUCAGUCCCCUCUACCCAUAUUCCUGCACUUAUCCUGGUUAUCCGAAUUCGGG .((((((.....(((.......((((((......))))))....((((((...))))))..)))))))))...........(((..((((....(((......))).....))))..))) (-24.14 = -24.70 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:27 2006