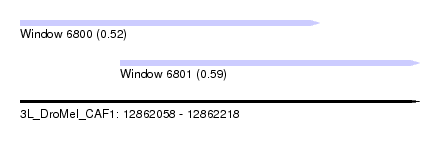
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,862,058 – 12,862,218 |
| Length | 160 |
| Max. P | 0.585009 |

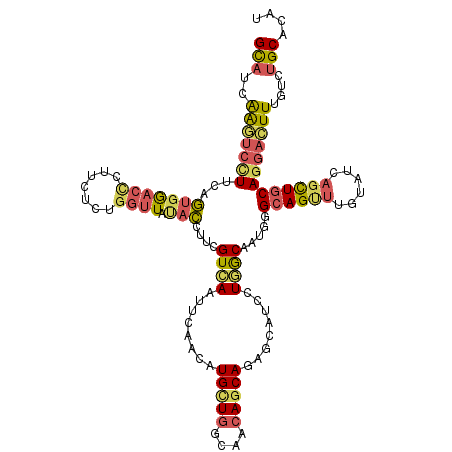
| Location | 12,862,058 – 12,862,178 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -32.61 |
| Consensus MFE | -21.36 |
| Energy contribution | -20.76 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12862058 120 + 23771897 GUAGUUCAGCUAAAACAUGGAGAAGCCCUUGAGUUCCACUGCAUCAAUUCUUUCAGUGGACCCUUCUCUGGUUAUACCUUCGUCAAUUCAACAUGCUGGCAACAGCAGAGCAUCCUGGCA ...((((.........((((((.((((...(((.(((((((.(........).))))))).....))).))))....))))))..........(((((....)))))))))......... ( -31.40) >DroSec_CAF1 100460 120 + 1 UUAGUUCAGCUAAAACAUGGAGAAGCCCUUGAGUUCCACUGCAUCAAGUCCUUCAGUGGACCCUUCUCUGGUUAUACCUUUGUCAAUUCAACAUGCUGGCAACAGCAGAGCAUCCUGGCA ((((.....))))(((.((((((((...((((((......)).))))((((......)))).))))))))))).......(((((........(((((....)))))........))))) ( -31.99) >DroSim_CAF1 101821 120 + 1 GUAGUUCAGCUAAAACAUGGGGAAGCCCUUGAGUUCCACUGCAUCAAGUCCUUCAGUGGACUCUUCUCUGGUUAUACAUUCGUCAAUUCAACAUGCUGGCAACAGCAGAGCAUCCUGGCA (((..((((.........(((....)))..((((.((((((............))))))))))....))))...)))....((((........(((((....)))))........)))). ( -35.89) >DroEre_CAF1 105849 120 + 1 AUAGUUCAGUUAAAACAUGGAGAGGCCCUAGAGUUUCACUGCAUCAAGUCCUUCAGUGGACUUUUCUCUGGUUAUACCUUCGUGAAUGCAACAUGCUGGCAACAGCAGAGUUUCCUCACA ...............(((((((.(...((((((............((((((......))))))..)))))).....)))))))).........(((((....)))))(((....)))... ( -31.44) >DroWil_CAF1 135956 120 + 1 UUAGUGGAAUUGAAACAUGGUGAGGCCAUGGAAUUACAUUGUCUUGGUGCGUUCAUUCAGCCAUUUGCCACAUAUCGACAAGUGACUGUCAGCUGCUGGCAGAAACAGAUCUUCCUCGCC ..................(((((((....(((.((((.(((((...(((.((..............))))).....)))))))))(((((((...))))))).......))).))))))) ( -29.24) >DroYak_CAF1 111848 120 + 1 AUAGUUCAGUUAAAACAUGGAGAGGCCCUAGAGUUUCACUGCAUCAAGUCCUUCAGUGGACUCUUCUCGGGUUACACCUUCGUGAAUUCAACAUGUUGGCAACAGCAGAGCUUCCUGACA ..(((((........(((((((.(((((.((((((.(((((............)))))))))))....)))))....))))))).........(((((....))))))))))........ ( -35.70) >consensus AUAGUUCAGCUAAAACAUGGAGAAGCCCUUGAGUUCCACUGCAUCAAGUCCUUCAGUGGACCCUUCUCUGGUUAUACCUUCGUCAAUUCAACAUGCUGGCAACAGCAGAGCAUCCUGGCA .............(((.((((((((.....((((......)).)).(((((......))))))))))))))))........((((........(((((....)))))........)))). (-21.36 = -20.76 + -0.60)

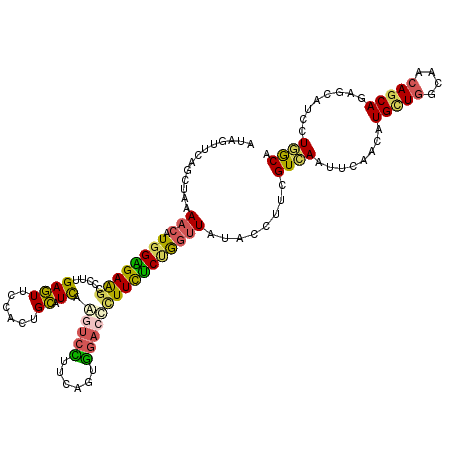

| Location | 12,862,098 – 12,862,218 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -23.17 |
| Energy contribution | -23.49 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12862098 120 + 23771897 GCAUCAAUUCUUUCAGUGGACCCUUCUCUGGUUAUACCUUCGUCAAUUCAACAUGCUGGCAACAGCAGAGCAUCCUGGCAAUGGGCAGUUUGUAUCAGCUGCAGGACUUUGUCUGCACUU ...............(..(((((......)).....(((..((((........(((((....)))))........)))).....((((((......))))))))).....)))..).... ( -31.09) >DroSec_CAF1 100500 120 + 1 GCAUCAAGUCCUUCAGUGGACCCUUCUCUGGUUAUACCUUUGUCAAUUCAACAUGCUGGCAACAGCAGAGCAUCCUGGCAAUGGGCAGCUUGUAUCAGCUGCAGGACUUUGUCUGCACUU (((.((((((((((((.(((...))).)))).....((.((((((........(((((....)))))........)))))).))((((((......))))))))))).)))..))).... ( -42.09) >DroSim_CAF1 101861 120 + 1 GCAUCAAGUCCUUCAGUGGACUCUUCUCUGGUUAUACAUUCGUCAAUUCAACAUGCUGGCAACAGCAGAGCAUCCUGGCAAUGGGCAGUUUGUAUCAGCUGCAGGACUUUGUCUGCACUU (((.((((((((((((.(((...))).)))).....((((.((((........(((((....)))))........)))))))).((((((......))))))))))).)))..))).... ( -36.89) >DroEre_CAF1 105889 120 + 1 GCAUCAAGUCCUUCAGUGGACUUUUCUCUGGUUAUACCUUCGUGAAUGCAACAUGCUGGCAACAGCAGAGUUUCCUCACAAUGGGCAGUUUGUACCAGCUGCAGGACUUUGUCUGCACAU (((.((((((((((((.(((...))).)))).....((...((((..(.(((.(((((....)))))..))).).))))...))((((((......))))))))))).)))..))).... ( -37.60) >DroWil_CAF1 135996 120 + 1 GUCUUGGUGCGUUCAUUCAGCCAUUUGCCACAUAUCGACAAGUGACUGUCAGCUGCUGGCAGAAACAGAUCUUCCUCGCCUUCGGUAGUCUUUAUAAUUUCCAGAAGUUUGUGUGCUCAU ....((((..(......).))))...((((((....((((......))))((((.((((.......(((.((.((........)).))))).........)))).)))))))).)).... ( -22.69) >DroYak_CAF1 111888 120 + 1 GCAUCAAGUCCUUCAGUGGACUCUUCUCGGGUUACACCUUCGUGAAUUCAACAUGUUGGCAACAGCAGAGCUUCCUGACAAUGGGCAGUUUGUAUCAGCUGCAGGACUUCGUCUGCACAU (((.((((((((((((.((((((.....(((((.(((....))))))))....(((((....)))))))).)))))))......((((((......))))))))))))).)..))).... ( -38.90) >consensus GCAUCAAGUCCUUCAGUGGACCCUUCUCUGGUUAUACCUUCGUCAAUUCAACAUGCUGGCAACAGCAGAGCAUCCUGGCAAUGGGCAGUUUGUAUCAGCUGCAGGACUUUGUCUGCACAU (((..(((((((...(((((((.......)))).)))....((((........(((((....)))))........)))).....((((((......)))))))))))))....))).... (-23.17 = -23.49 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:22 2006