
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,861,549 – 12,861,667 |
| Length | 118 |
| Max. P | 0.931078 |

| Location | 12,861,549 – 12,861,667 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -41.04 |
| Consensus MFE | -31.97 |
| Energy contribution | -33.11 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12861549 118 - 23771897 UUAGCUACUGGCAGUUGUUUGGUCUCGGCAUUCAGAUACAGAGGUGUCUGGACACCUUGUGAAUUGGGUAGAGUUGAGAUAUCAUAGCUGCCGGCAUGUACACUCACAUUAAGUGCCU .........(((((((((...((((((((.(((((((((....))))))))).((((........))))...))))))))...)))))))))(((((...............))))). ( -44.76) >DroSec_CAF1 99951 118 - 1 UUAGCUACUGGCAGUUGUUUGGUCUCAGCAUUCAGAUACAAAGGUGUCUGGACACCUUGUGAAUUGGGUAGAGUUGAGAUAUCAUAGCUGCCGGCAUGUACACUCACAUUUAGUGCCU .........(((((((((...((((((((.(((((((((....))))))))).((((........))))...))))))))...)))))))))(((((...............))))). ( -45.56) >DroSim_CAF1 101312 118 - 1 UUAGCUACUGGCAGUUGUUUGGUCUCAGCAUUCAGAUACAAAGGUGUCUGGACACCUUGUGAAUUGGGUAGAGUUGAGAUAUCAUAGCUGCCGGCAUGUACACUCACAUUUAGUGCCU .........(((((((((...((((((((.(((((((((....))))))))).((((........))))...))))))))...)))))))))(((((...............))))). ( -45.56) >DroEre_CAF1 105340 118 - 1 UUAGCUACUGGCAGUUGUUUGGUCUCGGCAUUUAGAUAAAGAGGUGUCUGGACACCUUGUGAAUUGGGCAGAGUUGAAACGUCAUAGCUGCCGGCAUGUACACUCAAAUUUAGAGCCU ...((..(((((((((((((.(.(((.((...........(((((((....))))))).........)).))).).))))).....))))))))...))...(((.......)))... ( -32.55) >DroWil_CAF1 135447 117 - 1 GUGG-CACUGGUAAUUGAUCAGUAUUAUGAUUUAAAUAUAGUUCCACUUGUUCACCAGCUGCAUUCGGUAGGGUGGAAACACCCCAAGCUCCUGCAUAGACACUUACAUUCAGUGCUU ((((-.((((......(((((......)))))......)))).)))).............(((((..((((((((....)))))...((....)).........)))....))))).. ( -32.20) >DroYak_CAF1 111339 118 - 1 UUAGCUACCGGCAGUUGUUUGGUCUCGGCAUUCAGGUAAAGAGGUGUCUGAACACCUUGUGAAUUGGGUAGAGUUGAGACAUCAUAGCUGCCGGCAUGUACACUCACAUUUAGUGCCU ...(((((((((((((((...((((((((((((((.....(((((((....))))))).....))))))...))))))))...))))))))))).((((......)))).)))).... ( -45.60) >consensus UUAGCUACUGGCAGUUGUUUGGUCUCAGCAUUCAGAUACAGAGGUGUCUGGACACCUUGUGAAUUGGGUAGAGUUGAGACAUCAUAGCUGCCGGCAUGUACACUCACAUUUAGUGCCU .........(((((((((...((((((((.((((((((......)))))))).((((........))))...))))))))...)))))))))(((((...............))))). (-31.97 = -33.11 + 1.14)

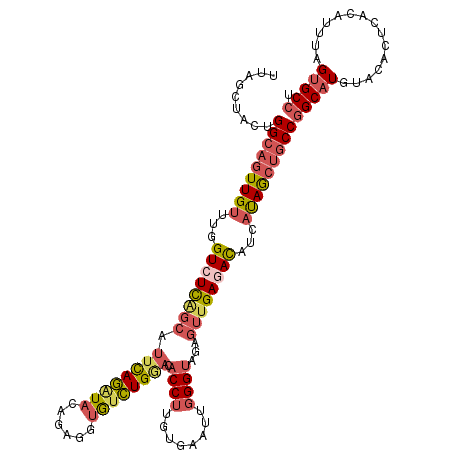
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:20 2006