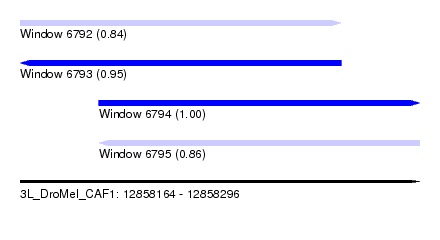
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,858,164 – 12,858,296 |
| Length | 132 |
| Max. P | 0.999176 |

| Location | 12,858,164 – 12,858,270 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 94.91 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -26.68 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12858164 106 + 23771897 GGCUGACCACUCGAUGUUGGCCAUGAGUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGCUUCCGUUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUU (((..((........))..)))....((((...)))).......(((((..((((((((.(((((..(((....)))...).)))).))))))))..))).))... ( -32.20) >DroSec_CAF1 96602 106 + 1 GGCUGACCACUCGGUAGUGGCCAUCAGUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGCUUCCGUUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUU (((.(((((..(((.((((..(((...((((((.((((......)).)).)))))).)))..)))).)))....))))).)))...(((((.........))))). ( -32.30) >DroSim_CAF1 97993 106 + 1 GGCUGACCACUCGGCAGUGGCCAUCAGUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGCUUCCGUUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUU (((.(((((..(((.((((..(((...((((((.((((......)).)).)))))).)))..)))).)))....))))).)))...(((((.........))))). ( -32.30) >DroEre_CAF1 101871 106 + 1 CCCUGACCACUCGGUAGUGGCCAUCAGUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGUUUCUGGGCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUU .......((((((((...(((.(((((....(((((((((((((((((...))))).))))))))...))))...((....)).)))))))).))))))))..... ( -30.00) >DroYak_CAF1 107993 106 + 1 GGCUGACCACUCGGUAGUGGCCAUCAGUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGUUGCUGGUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUU (((.(((((...(((....))).......(((((((((((((((((((...))))).))))))))))..)))).))))).)))...(((((.........))))). ( -36.50) >consensus GGCUGACCACUCGGUAGUGGCCAUCAGUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGCUUCCGUUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUU (((((((((((....)))))...))))))...............(((((..((((((((.(((((..(((....)))...).)))).))))))))..))).))... (-26.68 = -26.84 + 0.16)


| Location | 12,858,164 – 12,858,270 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 94.91 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -21.76 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12858164 106 - 23771897 AAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGAACGGAAGCAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAACUCAUGGCCAACAUCGAGUGGUCAGCC ..((((.........))))....(((.((((..((((....)..)))....(((((....((......))...)))))(((((((.....))).)))))))).))) ( -25.60) >DroSec_CAF1 96602 106 - 1 AAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGAACGGAAGCAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAACUGAUGGCCACUACCGAGUGGUCAGCC ..((((.........))))....(((.(((((....(((.((...((((..(((((....((......))...)))))....))))...)).)))..))))).))) ( -26.40) >DroSim_CAF1 97993 106 - 1 AAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGAACGGAAGCAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAACUGAUGGCCACUGCCGAGUGGUCAGCC ..((((.........))))....(((.(((((....(((..((..(((...(((((....((......))...)))))..)))..)).....)))..))))).))) ( -26.50) >DroEre_CAF1 101871 106 - 1 AAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGCCCAGAAACAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAACUGAUGGCCACUACCGAGUGGUCAGGG .......(((.(((((.......(((.......)))...............(((((....((......))...))))).)))))(((((((....))))))).))) ( -26.20) >DroYak_CAF1 107993 106 - 1 AAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGACCAGCAACAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAACUGAUGGCCACUACCGAGUGGUCAGCC ..((((.........))))....(((.......((((..(((((((.((..(((.....))).)).)))))))))))......((((((((....))))))))))) ( -28.30) >consensus AAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGAACGGAAGCAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAACUGAUGGCCACUACCGAGUGGUCAGCC ..((((.........))))....(((.((((.........((((((.((..(((.....))).)).)))))).))))......((((((((....))))))))))) (-21.76 = -22.32 + 0.56)

| Location | 12,858,190 – 12,858,296 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -25.76 |
| Energy contribution | -25.28 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.999176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
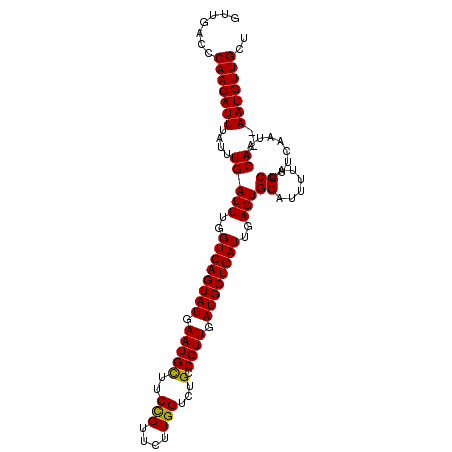
>3L_DroMel_CAF1 12858190 106 + 23771897 GUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGCUUCCGUUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUUUAGCUUUCAAUAACAAAAAUGUUGCU .......((((((((..((((((..((((((((.(((((..(((....)))...).)))).))))))))..)))((......)).........))))))))))).. ( -28.90) >DroSec_CAF1 96628 104 + 1 GUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGCUUCCGUUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUUUAGCUUUCAAUAACA--AAUGUUGCU .......((((((((..((((((..((((((((.(((((..(((....)))...).)))).))))))))..)))((......))...)))....)--))))))).. ( -28.60) >DroSim_CAF1 98019 104 + 1 GUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGCUUCCGUUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUUUAGCUUUCAAUAACA--AAUGUUGCU .......((((((((..((((((..((((((((.(((((..(((....)))...).)))).))))))))..)))((......))...)))....)--))))))).. ( -28.60) >DroEre_CAF1 101897 104 + 1 GUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGUUUCUGGGCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUUAAGCUUUCAAUAACA--AAUGUUGCU .......((((((((..((((((..((((((((.(((.......(((.......)))))).))))))))..)))((......))...)))....)--))))))).. ( -26.01) >DroYak_CAF1 108019 104 + 1 GUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGUUGCUGGUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUUUAGCUUUCAAUAACA--AAUGUUGUU ((((((((((((((((((((((...))))).))))))))))..))))..((....))(((((.(((((.((....))..)))))..)))))))).--......... ( -26.80) >consensus GUUGACCCAACAUUUAUUUGAUUUGGUCAGUAUGAAUGCUUCCGUUCUUGGUCUGCCGUUGAUGCUGAUUGAGUGCAUUUUAGCUUUCAAUAACA__AAUGUUGCU .......(((((((....(((((..((((((((.(((((..(((....)))...).)))).))))))))..)))((......)).........))..))))))).. (-25.76 = -25.28 + -0.48)

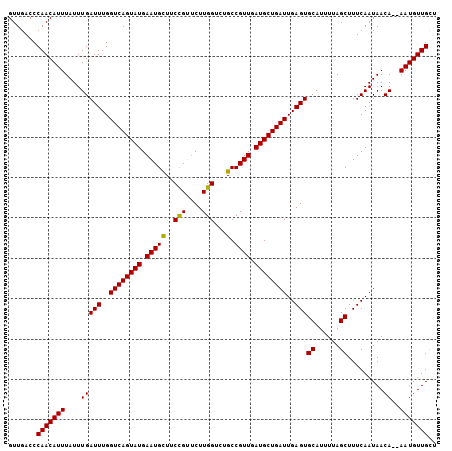
| Location | 12,858,190 – 12,858,296 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -20.39 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12858190 106 - 23771897 AGCAACAUUUUUGUUAUUGAAAGCUAAAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGAACGGAAGCAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAAC ..((((((((.(((((((((..((......))..))))).)))).....(((((........(....)........))).)).........))))))))....... ( -20.19) >DroSec_CAF1 96628 104 - 1 AGCAACAUU--UGUUAUUGAAAGCUAAAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGAACGGAAGCAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAAC ..(((((((--(((((((((..((......))..)))))..........(((((........(....)........))).))......)))))))))))....... ( -20.99) >DroSim_CAF1 98019 104 - 1 AGCAACAUU--UGUUAUUGAAAGCUAAAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGAACGGAAGCAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAAC ..(((((((--(((((((((..((......))..)))))..........(((((........(....)........))).))......)))))))))))....... ( -20.99) >DroEre_CAF1 101897 104 - 1 AGCAACAUU--UGUUAUUGAAAGCUUAAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGCCCAGAAACAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAAC ..(((((((--((((.((((..((......))..))))((((.......(((.......)))..((.....))...))))........)))))))))))....... ( -20.40) >DroYak_CAF1 108019 104 - 1 AACAACAUU--UGUUAUUGAAAGCUAAAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGACCAGCAACAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAAC .........--(((((((((..((......))..))))).)))).....((....))..((((..(((((((.((..(((.....))).)).)))))))))))... ( -19.40) >consensus AGCAACAUU__UGUUAUUGAAAGCUAAAAUGCACUCAAUCAGCAUCAACGGCAGACCAAGAACGGAAGCAUUCAUACUGACCAAAUCAAAUAAAUGUUGGGUCAAC ..(((((((..(((((((((..((......))..))))).)))).....(((((......................))).))..........)))))))....... (-16.37 = -16.37 + -0.00)


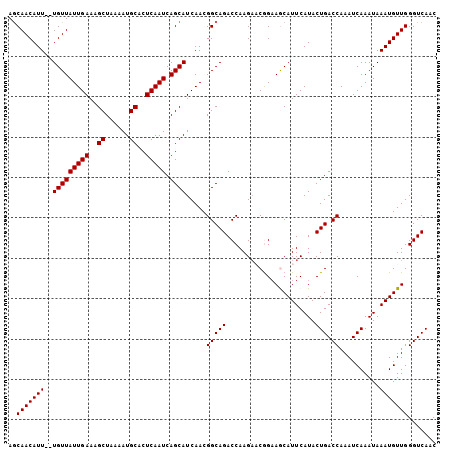
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:16 2006