
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,856,963 – 12,857,207 |
| Length | 244 |
| Max. P | 0.895705 |

| Location | 12,856,963 – 12,857,079 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -24.34 |
| Energy contribution | -25.58 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12856963 116 - 23771897 UGAUUGCAGUUACUUAAUGAAAGUGCAUAAUUCGACAGGGUUACUGCAGUGAAGGCAGCAGAUGCUUAGGACGAGCAUUGCU-GGGUGGAAAUUAUUGAUUUUCCGAUAGUUUGCUG (.(((((((.(((((.....)))))..((((((....))))))))))))).)...((((((.(((((.....))))))))))-)...(.(((((((((......))))))))).).. ( -32.90) >DroSec_CAF1 95341 116 - 1 UGAUUGCAGUAACUUAAUGAAAGUGCAUAAUUCGACAGGGUCACUGCAGUCAUGGCAGCAGAUGCUUAGAACGAGCAUAGCU-GGGUGGAAAUUAUUGAUUUUCAGAUAGUUUGCUG ((((((((((.((((..((((.........))))...)))).))))))))))...((((((((((((.....)))))).(((-(..((((((((...))))))))..)))))))))) ( -35.80) >DroSim_CAF1 96823 116 - 1 UGAUUGCAGUUACUUAAUGAAAGUGCAUAAUUCGACAGGGUCACUGCAGUCAUGGCAGCAGAUGCUUAGGACGAGCAAUGCU-GGGUGAAGAUUAUUGAUUUUCAGAUAGUUUGCUG ((((((((((.((((..((((.........))))...)))).))))))))))...((((((.(((((.....)))))..(((-(..((((((((...))))))))..)))))))))) ( -35.20) >DroEre_CAF1 100659 115 - 1 UGAUUGCAGUUACUUAAUGAAAGUGCAUAAUUCGAUAGGGUCACUGCAGUCAAGGCAGCAGAUGCUUAGGACAAGGAUUGCU-GGGAGGAAAUUAUUGAUUUUCU-AUAGGUUGCUG ((((((((((..((((.((((.........)))).))))...)))))))))).((((((.(((.(((.....))).))).((-(..((((((((...))))))))-.))))))))). ( -33.20) >DroYak_CAF1 106774 117 - 1 UGAUUGCAGUUAGUUAAUGAAAGUGCAUAAUUCGAUAGGGUCACUGCAGUCAAGGCAACAGAUGCUUAGGACAAGGAUUGCUUGGGGGGAAACUAUUGAUUUUCGGAUAGUUUGCUG ((((((((((......(((......))).((((....)))).))))))))))(((((.....)))))....((((.....))))..((.((((((((........)))))))).)). ( -27.80) >consensus UGAUUGCAGUUACUUAAUGAAAGUGCAUAAUUCGACAGGGUCACUGCAGUCAAGGCAGCAGAUGCUUAGGACGAGCAUUGCU_GGGUGGAAAUUAUUGAUUUUCAGAUAGUUUGCUG ((((((((((.((((..((((.........))))...)))).)))))))))).((((((((.(((((.....)))))))))).......((((((((........))))))))))). (-24.34 = -25.58 + 1.24)



| Location | 12,857,002 – 12,857,103 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.18 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12857002 101 + 23771897 UGCUCGUCCUAAGCAUCUGCUGCCUUCACUGCAGUAACCCUGUCGAAUUAUGCACUUUCAUUAAGUAACUGCAAUCAGGCGAAUAAAAAUCCUUUCGAGAG ..((((.....(((....)))((((....((((((.((...((.(((.........)))))...)).))))))...))))...............)))).. ( -21.00) >DroSec_CAF1 95380 101 + 1 UGCUCGUUCUAAGCAUCUGCUGCCAUGACUGCAGUGACCCUGUCGAAUUAUGCACUUUCAUUAAGUUACUGCAAUCAGGCGAAUAAAAAUCCUUUCGAGAG ..((((.....(((....)))(((.(((.(((((((((...((.(((.........)))))...))))))))).))))))...............)))).. ( -28.10) >DroSim_CAF1 96862 101 + 1 UGCUCGUCCUAAGCAUCUGCUGCCAUGACUGCAGUGACCCUGUCGAAUUAUGCACUUUCAUUAAGUAACUGCAAUCAGGCGAAUAAAAAUCCUUUCGAGAG ..((((.....(((....)))(((.(((.(((((((((...))).........((((.....)))).)))))).))))))...............)))).. ( -25.30) >DroEre_CAF1 100697 101 + 1 UCCUUGUCCUAAGCAUCUGCUGCCUUGACUGCAGUGACCCUAUCGAAUUAUGCACUUUCAUUAAGUAACUGCAAUCAGGCGAAUAAAAAUCCUUUCGAGAG ..((((.....(((....)))((((.((.((((((.((..((..(((.........)))..)).)).)))))).))))))...............)))).. ( -21.90) >DroYak_CAF1 106814 101 + 1 UCCUUGUCCUAAGCAUCUGUUGCCUUGACUGCAGUGACCCUAUCGAAUUAUGCACUUUCAUUAACUAACUGCAAUCAGGCGAAUAAAAAUCCUUUCGAGAG ..((((.............((((((.((.((((((......((.(((.........)))))......)))))).)))))))).............)))).. ( -21.27) >consensus UGCUCGUCCUAAGCAUCUGCUGCCUUGACUGCAGUGACCCUGUCGAAUUAUGCACUUUCAUUAAGUAACUGCAAUCAGGCGAAUAAAAAUCCUUUCGAGAG ..((((.....(((....)))(((.(((.((((((.((...((.(((.........)))))...)).)))))).))))))...............)))).. (-21.42 = -21.18 + -0.24)

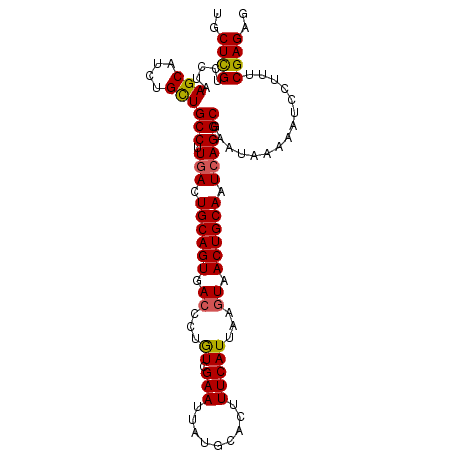

| Location | 12,857,002 – 12,857,103 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.46 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12857002 101 - 23771897 CUCUCGAAAGGAUUUUUAUUCGCCUGAUUGCAGUUACUUAAUGAAAGUGCAUAAUUCGACAGGGUUACUGCAGUGAAGGCAGCAGAUGCUUAGGACGAGCA ..((((...((((....))))((((.(((((((.(((((.....)))))..((((((....)))))))))))))..))))(((....))).....)))).. ( -27.10) >DroSec_CAF1 95380 101 - 1 CUCUCGAAAGGAUUUUUAUUCGCCUGAUUGCAGUAACUUAAUGAAAGUGCAUAAUUCGACAGGGUCACUGCAGUCAUGGCAGCAGAUGCUUAGAACGAGCA ..((((...((((....))))(((((((((((((.((((..((((.........))))...)))).)))))))))).)))(((....))).....)))).. ( -30.00) >DroSim_CAF1 96862 101 - 1 CUCUCGAAAGGAUUUUUAUUCGCCUGAUUGCAGUUACUUAAUGAAAGUGCAUAAUUCGACAGGGUCACUGCAGUCAUGGCAGCAGAUGCUUAGGACGAGCA ..((((...((((....))))(((((((((((((.((((..((((.........))))...)))).)))))))))).)))(((....))).....)))).. ( -30.00) >DroEre_CAF1 100697 101 - 1 CUCUCGAAAGGAUUUUUAUUCGCCUGAUUGCAGUUACUUAAUGAAAGUGCAUAAUUCGAUAGGGUCACUGCAGUCAAGGCAGCAGAUGCUUAGGACAAGGA .((.(....)))(((((.((((((((((((((((..((((.((((.........)))).))))...))))))))).))))(((....)))..))).))))) ( -29.20) >DroYak_CAF1 106814 101 - 1 CUCUCGAAAGGAUUUUUAUUCGCCUGAUUGCAGUUAGUUAAUGAAAGUGCAUAAUUCGAUAGGGUCACUGCAGUCAAGGCAACAGAUGCUUAGGACAAGGA .(((..(..((((....))))(((((((((((((......(((......))).((((....)))).))))))))).)))).........)..)))...... ( -22.10) >consensus CUCUCGAAAGGAUUUUUAUUCGCCUGAUUGCAGUUACUUAAUGAAAGUGCAUAAUUCGACAGGGUCACUGCAGUCAAGGCAGCAGAUGCUUAGGACGAGCA ..((((...((((....))))(((((((((((((.((((..((((.........))))...)))).)))))))))).)))...............)))).. (-24.66 = -25.46 + 0.80)


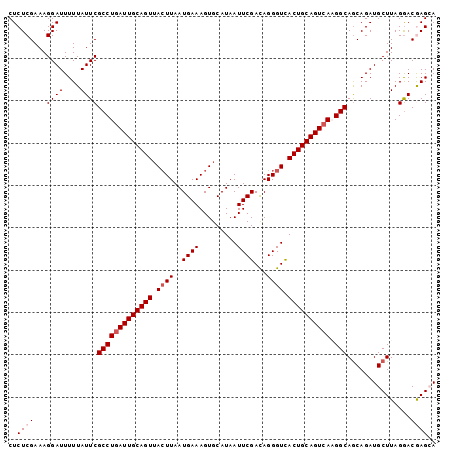
| Location | 12,857,103 – 12,857,207 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 94.40 |
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -30.02 |
| Energy contribution | -30.18 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12857103 104 + 23771897 CAAUCUGUUGGCUUGGCUGGCUGCCUUAACCUUGUGACUGGCUGAGCGUGGCUUAAUGCACUUUUAAUUGAGCCAAGCAUCCUGGGCUAGCUAGGGGGAAAUUG ...........((((((((((((((..............)))...((.(((((((((.........))))))))).))......)))))))))))......... ( -34.84) >DroSec_CAF1 95481 104 + 1 CAAUCUGUUGGCUUGGCUGGCUGCCGUAACCUUGUGACUGGCUGAGCGUGGCUUAAUGCACUUUUAAUUGAGCCAAGCAUCCUGGGCUAGCGAGCGGGAAAUUG ((((...((.(((((.(((((((((((.((...)).)).)))...((.(((((((((.........))))))))).))......))))))))))).))..)))) ( -35.40) >DroSim_CAF1 96963 104 + 1 CAAUCUGUUGGCUUGGCUGGCUGCCUUAACCUUGUGACUGGCUGAGCGUGGCUUAAUGCACUUUUAAUUGAGCCGAGCAUCCUGUUCUAGCGAGCGGGAAAUUG .....((((.(((..((..(..((.........))..)..))..))).(((((((((.........)))))))))))))((((((((....))))))))..... ( -37.40) >DroEre_CAF1 100798 100 + 1 CAAUCUGUUGGCUUGGCUGGCUGCCUUAACCUUCUGACUGGCUGAGCGUGGCUUAAUGCACUUUUAAUUGAGCCGAGCAUCCUGGG----CGAGCGGGAAAUUG ...((((((.(((..(...(((((((((.((........)).)))).))((((((((.........)))))))).)))...)..))----).))))))...... ( -34.50) >DroYak_CAF1 106915 100 + 1 CAAUCUGUUGGCUUGGCUGGCUGCCUUAACCUUGUGACUGGCUGAGCGUGGCUUAAUGCACUUUUAAUUGAGCCGAGCAUCCUGGG----CGGGCGGGAAAUUG ...((((((.(((..(...(((((((((.((........)).)))).))((((((((.........)))))))).)))...)..))----).))))))...... ( -34.40) >consensus CAAUCUGUUGGCUUGGCUGGCUGCCUUAACCUUGUGACUGGCUGAGCGUGGCUUAAUGCACUUUUAAUUGAGCCGAGCAUCCUGGGCUAGCGAGCGGGAAAUUG .....((((.(((..((..(..((.........))..)..))..))).(((((((((.........)))))))))))))(((((..........)))))..... (-30.02 = -30.18 + 0.16)


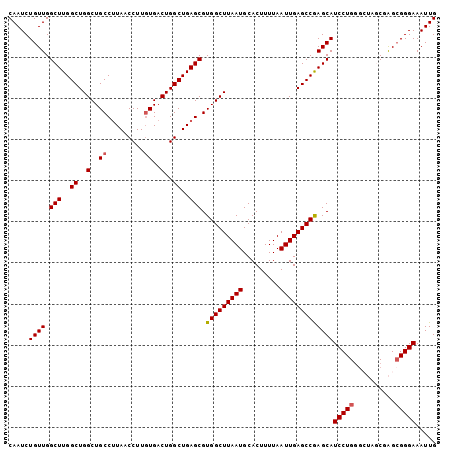
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:12 2006