
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,856,140 – 12,856,270 |
| Length | 130 |
| Max. P | 0.946221 |

| Location | 12,856,140 – 12,856,240 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.86 |
| Mean single sequence MFE | -18.63 |
| Consensus MFE | -18.15 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
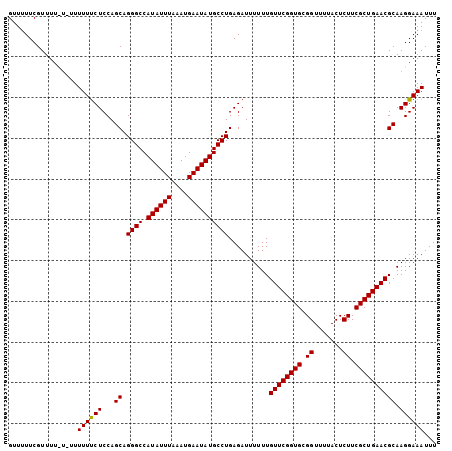
>3L_DroMel_CAF1 12856140 100 + 23771897 AAAUUUCCUUGCGUUCAGCGAAGAGUAAAACCGCACCGAACAAAAAAUCUCAGGCAUAUUCAUUUAAAUAUGGCCCUGCUGGAGAAAAA----AAAACGAA-AAC ...(((((..((((((.(((...........)))...))))...........((((((((......))))).)))..)).)))))....----........-... ( -18.20) >DroSec_CAF1 94446 104 + 1 AAAUUUCCUUGCGUUCAGCGAAGAGUAAAACCGCACCGAACAAAAAAUCUCAGGCAUAUUCAUUUAAAUAUGGCCCUGCUGGAGGAAAAA-AGAAAACGAAAAAC ...(((((((..((((.(((...........)))...)))).........((((((((((......))))))..))))...)))))))..-.............. ( -19.50) >DroSim_CAF1 95951 105 + 1 AAAUUUCCUUGCGUUCAGCGAAGAGUAAAACCGCACCGAACAAAAAAUCUCAGGCAUAUUCAUUUAAAUAUGGCCCUGCUGGAGAAAAAAAAAAAAACGAAAAAC ...(((((..((((((.(((...........)))...))))...........((((((((......))))).)))..)).))))).................... ( -18.20) >consensus AAAUUUCCUUGCGUUCAGCGAAGAGUAAAACCGCACCGAACAAAAAAUCUCAGGCAUAUUCAUUUAAAUAUGGCCCUGCUGGAGAAAAAA_A_AAAACGAAAAAC ...(((((..((((((.(((...........)))...))))...........((((((((......))))).)))..)).))))).................... (-18.15 = -17.93 + -0.22)



| Location | 12,856,140 – 12,856,240 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.86 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -24.99 |
| Energy contribution | -24.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
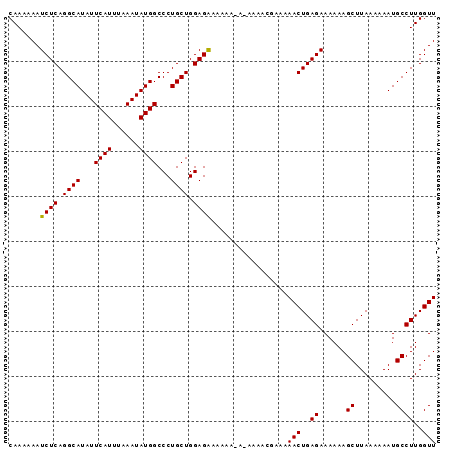
>3L_DroMel_CAF1 12856140 100 - 23771897 GUU-UUCGUUUU----UUUUUCUCCAGCAGGGCCAUAUUUAAAUGAAUAUGCCUGAGAUUUUUUGUUCGGUGCGGUUUUACUCUUCGCUGAACGCAAGGAAAUUU (((-(((.....----....((((.....((((.((((((....))))))))))))))......((((((((.((.......)).))))))))....)))))).. ( -24.50) >DroSec_CAF1 94446 104 - 1 GUUUUUCGUUUUCU-UUUUUCCUCCAGCAGGGCCAUAUUUAAAUGAAUAUGCCUGAGAUUUUUUGUUCGGUGCGGUUUUACUCUUCGCUGAACGCAAGGAAAUUU ..............-..((((((...((.((((.((((((....))))))))))..........((((((((.((.......)).)))))))))).))))))... ( -26.70) >DroSim_CAF1 95951 105 - 1 GUUUUUCGUUUUUUUUUUUUUCUCCAGCAGGGCCAUAUUUAAAUGAAUAUGCCUGAGAUUUUUUGUUCGGUGCGGUUUUACUCUUCGCUGAACGCAAGGAAAUUU ......................(((.((.((((.((((((....))))))))))..........((((((((.((.......)).))))))))))..)))..... ( -23.90) >consensus GUUUUUCGUUUU_U_UUUUUUCUCCAGCAGGGCCAUAUUUAAAUGAAUAUGCCUGAGAUUUUUUGUUCGGUGCGGUUUUACUCUUCGCUGAACGCAAGGAAAUUU .................((((((...((.((((.((((((....))))))))))..........((((((((.((.......)).)))))))))).))))))... (-24.99 = -24.77 + -0.22)


| Location | 12,856,180 – 12,856,270 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -13.77 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.99 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
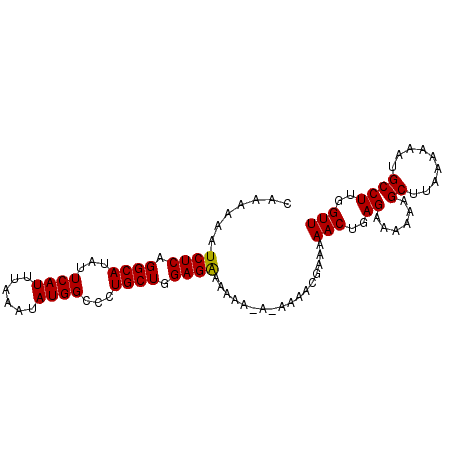
>3L_DroMel_CAF1 12856180 90 + 23771897 CAAAAAAUCUCAGGCAUAUUCAUUUAAAUAUGGCCCUGCUGGAGAAAAA----AAAACGAA-AACUGAGAAAAAAGCUUAAAAAAUGCCUUGGUU .......((((.((((...((((......))))...)))).))))....----........-(((..((......((.........))))..))) ( -13.80) >DroSec_CAF1 94486 94 + 1 CAAAAAAUCUCAGGCAUAUUCAUUUAAAUAUGGCCCUGCUGGAGGAAAAA-AGAAAACGAAAAACUGAGAAAAAAGCUUAAAAAAUGCCUUGGUU (((....((((((.((((((......))))))..(((.....))).....-.............)))))).....((.........)).)))... ( -13.70) >DroSim_CAF1 95991 95 + 1 CAAAAAAUCUCAGGCAUAUUCAUUUAAAUAUGGCCCUGCUGGAGAAAAAAAAAAAAACGAAAAACUGAGAAAAAAGCUUAAAAAAUGCCUUGGUU .......((((.((((...((((......))))...)))).)))).................(((..((......((.........))))..))) ( -13.80) >consensus CAAAAAAUCUCAGGCAUAUUCAUUUAAAUAUGGCCCUGCUGGAGAAAAAA_A_AAAACGAAAAACUGAGAAAAAAGCUUAAAAAAUGCCUUGGUU .......((((.((((...((((......))))...)))).)))).................(((..((......((.........))))..))) (-13.62 = -13.40 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:08 2006