
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,855,866 – 12,856,060 |
| Length | 194 |
| Max. P | 0.992108 |

| Location | 12,855,866 – 12,855,984 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.45 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.66 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12855866 118 + 23771897 CUUUCUUAGGUUGCACAUAAAAGCGACAACUUCUGCGGCAAAACUCGCUUAAUUUGCAUGAAAUAAGUUUUAGGCCAGGAAACUA--UCUCUUGGCCCAGAGACUUGAAGUCUGUGGAUA .........(((((........))))).......((((......))))...(((..((.((..((((((((.((((((((.....--..))))))))..))))))))...))))..))). ( -34.50) >DroSec_CAF1 94166 120 + 1 CUUCCUAAGUUUGCAUAUAAAAGCGACAACUUCUGCGGCAAAACUCGCUUAAUUUGCAUGAAAAAAGUUUUAGGCCAGGAAACUAUAUCUCUUGGCCCAGAGACUUGAAGUCUGUGUACU ......((((((((........))))..))))..((((......))))......(((((((...(((((((.((((((((.........))))))))..)))))))....)).))))).. ( -28.60) >DroSim_CAF1 95671 120 + 1 CUUCCUAAGUUUGCACAUAAAAGCGAAAACUUCUGCGGCAAAACUCGCUUAAUUUGCAUGAAAUAAGUUUUAGGCCAGGAAACUAUAUAUCUUGGCCCAGAGACUUGAAGUCUGUGGACU .......((((..((.......(((((.......((((......))))....)))))..((..((((((((.((((((((.........))))))))..))))))))...))))..)))) ( -32.40) >DroEre_CAF1 99576 117 + 1 CUUUCUUAGUUUGCACAUAAAAGCCACAACUUCUCCGGCA-AACUCUCUUUAUUUGCAUAAAAUAAGUUUUAGGCCAGGAAACUA--UCUCUUGGCCCAGAGACAUGAAGUCUGUGGACU .....................((((((((((((....(((-((.........))))).........(((((.((((((((.....--..))))))))..)))))..))))).))))).)) ( -31.40) >DroYak_CAF1 105709 118 + 1 CUUUCUUAGCUUGCACAUAAAUGCCGCAACUUCUGCUGCAAAACUCUCUUCAUUUGCAUAAAAUAAGUUUUAGGCACGGAAACUA--UCUCUUGGUCCAAAGACAUGAAGUCUGUGGACU .....................(((((((.....)))((((((..........))))))..............)))).(....)..--......((((((.((((.....)))).)))))) ( -25.70) >consensus CUUUCUUAGUUUGCACAUAAAAGCGACAACUUCUGCGGCAAAACUCGCUUAAUUUGCAUGAAAUAAGUUUUAGGCCAGGAAACUA__UCUCUUGGCCCAGAGACUUGAAGUCUGUGGACU ............((........)).((((((((....(((((..........))))).......(((((((.((((((((.........))))))))..)))))))))))).)))..... (-21.94 = -22.66 + 0.72)

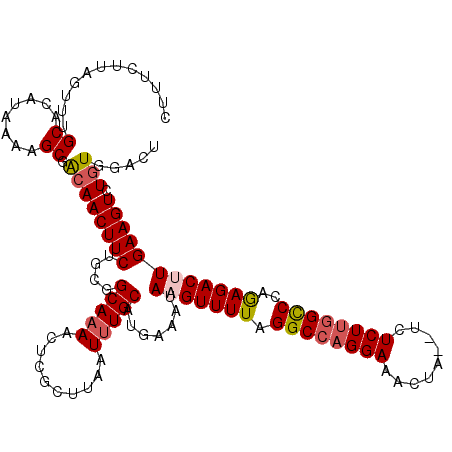

| Location | 12,855,906 – 12,856,020 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.93 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -26.29 |
| Energy contribution | -26.45 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12855906 114 + 23771897 AAACUCGCUUAAUUUGCAUGAAAUAAGUUUUAGGCCAGGAAACUA--UCUCUUGGCCCAGAGACUUGAAGUCUGUGGAUAUCACUUCCGCGUUGCCCAUAGAGUAUA----UGUUCAAAU ..((((((.......))(((...((((((((.((((((((.....--..))))))))..))))))))......(((((.......)))))......))).))))...----......... ( -31.90) >DroSec_CAF1 94206 120 + 1 AAACUCGCUUAAUUUGCAUGAAAAAAGUUUUAGGCCAGGAAACUAUAUCUCUUGGCCCAGAGACUUGAAGUCUGUGUACUUCACUUCCGCGUUGCCCAUAGAGUAUAUGUAUGUUCAAAU .....(((................(((((((.((((((((.........))))))))..)))))))(((((......)))))......)))(((..((((.(.....).))))..))).. ( -28.50) >DroSim_CAF1 95711 120 + 1 AAACUCGCUUAAUUUGCAUGAAAUAAGUUUUAGGCCAGGAAACUAUAUAUCUUGGCCCAGAGACUUGAAGUCUGUGGACUUCACUUCCGCGUUGCCCACAGAGUAUAUGUAUGUUCAAAU ..........(((.((((((...((((((((.((((((((.........))))))))..)))))))).(.(((((((.(..(.(....).)..).))))))).).)))))).)))..... ( -32.60) >DroEre_CAF1 99616 113 + 1 -AACUCUCUUUAUUUGCAUAAAAUAAGUUUUAGGCCAGGAAACUA--UCUCUUGGCCCAGAGACAUGAAGUCUGUGGACUUCACUUCCGCGUUGCCCAUAGAGUAUG----UGUUCAGAU -.((...((((((..(((........(((((.((((((((.....--..))))))))..))))).(((((((....))))))).........)))..))))))...)----)........ ( -32.40) >DroYak_CAF1 105749 114 + 1 AAACUCUCUUCAUUUGCAUAAAAUAAGUUUUAGGCACGGAAACUA--UCUCUUGGUCCAAAGACAUGAAGUCUGUGGACUUCACUUCCGUGUUGCCCAUAGAGUAUA----UGUUCAGAU ....(((...(((.(((.(((((....)))))(((((((((....--......((((((.((((.....)))).))))))....))))))))).........))).)----))...))). ( -28.04) >consensus AAACUCGCUUAAUUUGCAUGAAAUAAGUUUUAGGCCAGGAAACUA__UCUCUUGGCCCAGAGACUUGAAGUCUGUGGACUUCACUUCCGCGUUGCCCAUAGAGUAUA____UGUUCAAAU ..((((........(((.........(((((.((((((((.........))))))))..))))).(((((((....))))))).....))).........))))................ (-26.29 = -26.45 + 0.16)


| Location | 12,855,906 – 12,856,020 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.93 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -26.12 |
| Energy contribution | -27.16 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12855906 114 - 23771897 AUUUGAACA----UAUACUCUAUGGGCAACGCGGAAGUGAUAUCCACAGACUUCAAGUCUCUGGGCCAAGAGA--UAGUUUCCUGGCCUAAAACUUAUUUCAUGCAAAUUAAGCGAGUUU .........----...((((....(....)((((((((((.......((((.....)))).(((((((.((((--...)))).)))))))....))))))).))).........)))).. ( -28.00) >DroSec_CAF1 94206 120 - 1 AUUUGAACAUACAUAUACUCUAUGGGCAACGCGGAAGUGAAGUACACAGACUUCAAGUCUCUGGGCCAAGAGAUAUAGUUUCCUGGCCUAAAACUUUUUUCAUGCAAAUUAAGCGAGUUU ..(((..((((.........))))..)))((((((..((((((......))))))..))).(((((((.(((((...))))).)))))))......................)))..... ( -31.10) >DroSim_CAF1 95711 120 - 1 AUUUGAACAUACAUAUACUCUGUGGGCAACGCGGAAGUGAAGUCCACAGACUUCAAGUCUCUGGGCCAAGAUAUAUAGUUUCCUGGCCUAAAACUUAUUUCAUGCAAAUUAAGCGAGUUU ....((((..........(((((((((..(((....)))..)))))))))..((((((...(((((((.((.........)).)))))))..)))).......((.......)))))))) ( -36.90) >DroEre_CAF1 99616 113 - 1 AUCUGAACA----CAUACUCUAUGGGCAACGCGGAAGUGAAGUCCACAGACUUCAUGUCUCUGGGCCAAGAGA--UAGUUUCCUGGCCUAAAACUUAUUUUAUGCAAAUAAAGAGAGUU- ....(((((----.....(((.(((((..(((....)))..))))).))).....))).))(((((((.((((--...)))).))))))).(((((.((((((....)))))).)))))- ( -34.10) >DroYak_CAF1 105749 114 - 1 AUCUGAACA----UAUACUCUAUGGGCAACACGGAAGUGAAGUCCACAGACUUCAUGUCUUUGGACCAAGAGA--UAGUUUCCGUGCCUAAAACUUAUUUUAUGCAAAUGAAGAGAGUUU .........----.........(((((...((((((((((((((....)))))))(((((((......)))))--)).))))))))))))((((((.((((((....)))))).)))))) ( -33.60) >consensus AUUUGAACA____UAUACUCUAUGGGCAACGCGGAAGUGAAGUCCACAGACUUCAAGUCUCUGGGCCAAGAGA__UAGUUUCCUGGCCUAAAACUUAUUUCAUGCAAAUUAAGCGAGUUU ................((((..(((((..(((....)))..))))).((((.....)))).(((((((.((((.....)))).)))))))........................)))).. (-26.12 = -27.16 + 1.04)


| Location | 12,855,946 – 12,856,060 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -29.66 |
| Energy contribution | -29.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12855946 114 - 23771897 GAGGAUAUGAGGUUGUCUCAUGUGUCGCGCUGUCAGUCAAAUUUGAACA----UAUACUCUAUGGGCAACGCGGAAGUGAUAUCCACAGACUUCAAGUCUCUGGGCCAAGAGA--UAGUU ..(((((((((.....))))))).))..((((((.............((----((.....))))(((..(((....)))....(((.((((.....)))).))))))....))--)))). ( -32.70) >DroSec_CAF1 94246 120 - 1 GAGGAUAUGAGGUUGUCUCAUGUGUCGCGCUGUCAGUCAAAUUUGAACAUACAUAUACUCUAUGGGCAACGCGGAAGUGAAGUACACAGACUUCAAGUCUCUGGGCCAAGAGAUAUAGUU ....(((((((.....))))))).(((((.((((..(((....))).....((((.....)))))))).)))))...((((((......)))))).((((((......))))))...... ( -31.60) >DroSim_CAF1 95751 120 - 1 GAGGAUAUGAGGUUGUCUCAUGUGUCGCGCUGUCAGUCAAAUUUGAACAUACAUAUACUCUGUGGGCAACGCGGAAGUGAAGUCCACAGACUUCAAGUCUCUGGGCCAAGAUAUAUAGUU (((.(((......))))))(((((((..(((..(((....(((((((...........(((((((((..(((....)))..))))))))).)))))))..))))))...))))))).... ( -36.30) >DroEre_CAF1 99655 114 - 1 GAGGAUAUGAGGUUGUCUCAUGUGUCGCGCUGUCAGUCAAAUCUGAACA----CAUACUCUAUGGGCAACGCGGAAGUGAAGUCCACAGACUUCAUGUCUCUGGGCCAAGAGA--UAGUU (((((((((((.....))))))).....(.((((((......))).)))----)....(((.(((((..(((....)))..))))).))))))).(((((((......)))))--))... ( -36.60) >DroYak_CAF1 105789 114 - 1 GAGGAUAUGAGGUUGUCUCAUGUGUCGCGCUGUCAGUCAAAUCUGAACA----UAUACUCUAUGGGCAACACGGAAGUGAAGUCCACAGACUUCAUGUCUUUGGACCAAGAGA--UAGUU ((((((((((((((.....((((((.......((((......)))))))----)))......(((((..(((....)))..)))))..))))))))))))))...........--..... ( -35.21) >consensus GAGGAUAUGAGGUUGUCUCAUGUGUCGCGCUGUCAGUCAAAUUUGAACA____UAUACUCUAUGGGCAACGCGGAAGUGAAGUCCACAGACUUCAAGUCUCUGGGCCAAGAGA__UAGUU ..(((((((((.....))))))).))..((((((((......))))...........((((...(((..(((....)))....(((.((((.....)))).)))))).))))...)))). (-29.66 = -29.86 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:05 2006