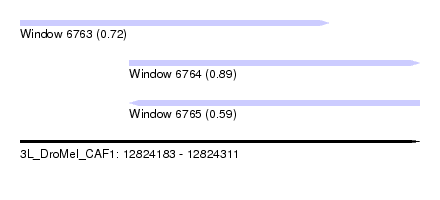
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,824,183 – 12,824,311 |
| Length | 128 |
| Max. P | 0.894472 |

| Location | 12,824,183 – 12,824,282 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.87 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12824183 99 + 23771897 AUUUUUGUGGACUCAAAACAGUUGGAAAAUUAAAGAGACAUGUCGGUUAAUACGGCUCACGCCAACAAUGGCGUGCUCAUCCAUGCUGGAGAGUAGUAA ..........((((....((((((((...............((((.......)))).(((((((....)))))))....)))).))))..))))..... ( -27.50) >DroVir_CAF1 81923 94 + 1 AUCUUUGUGUUACC-----UAUUGAAAAAUCUAUUCGGCAUGUCGGUCAACACGGCACACGCAAAUAAUGGUGUGCUCAUACAUGCGGGCGAGUAGUAA ..............-----...........(((((((((((((((.......))((((((.((.....))))))))....))))))...)))))))... ( -23.60) >DroSec_CAF1 65361 84 + 1 AUUUUUGUG---------------GAAAAUUAAAGAGACAUGUCGGUUAAUACGGCUCACGCCAACAACGGCGUGCUCAUCCAUGCUGGAGAGUAGUAA .........---------------..........(((.(((((((((....))(((....))).....)))))))))).(((.....)))......... ( -20.60) >DroYak_CAF1 69059 99 + 1 AUUUUUGUGGAUUCAAAACAGUUAAAAAAAUAAAGAGACAUGUCGGUUAAUACGGCUCACGCCAACAAUGGCGUGCUCAUCCACGCUGGAGAGUAGUAA ......((((((........(((.....)))...(((....((((.......)))).(((((((....))))))))))))))))((((.....)))).. ( -24.50) >DroMoj_CAF1 74766 94 + 1 AUCUUUGAGAAAAU-----UAUUUAAAAAACUAUCCAACAUGUCGAUCAAUACGGCACACGCAAAUAAUGGUGUGCUCAUACAUGCAGGAGAGUAGUAA ..............-----..........(((.(((..(((((((.......))((((((.((.....))))))))....)))))..))).)))..... ( -18.90) >DroAna_CAF1 60039 94 + 1 AUUUUGGUGGAC-----AUAGUUGGAUAAUCCGUUCAGCAUGUCGGUUAACACGGCUCACGCCAACAAUGGCGUGCUUAUCCACGCCGGAGAGUAGUAA .(((((((((((-----((.(((((((.....)))))))))))).........((..(((((((....))))))).....)).))))))))........ ( -32.50) >consensus AUUUUUGUGGAA_C_____AGUUGAAAAAUCAAAGAGACAUGUCGGUUAAUACGGCUCACGCCAACAAUGGCGUGCUCAUCCAUGCUGGAGAGUAGUAA ......(((((..............................((((.......)))).((((((......))))))....)))))............... (-14.28 = -14.83 + 0.56)

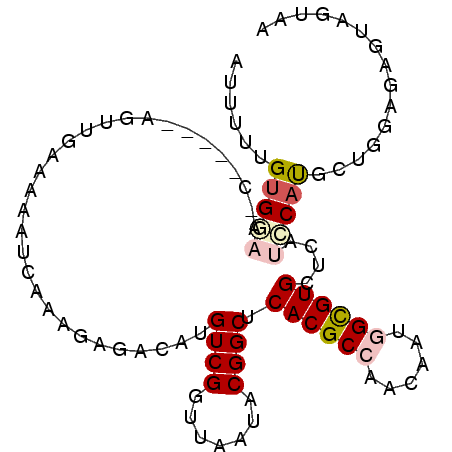
| Location | 12,824,218 – 12,824,311 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.55 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
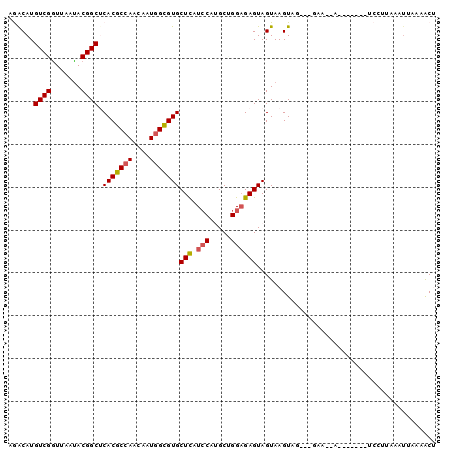
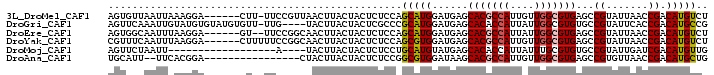
>3L_DroMel_CAF1 12824218 93 + 23771897 AGACAUGUCGGUUAAUACGGCUCACGCCAACAAUGGCGUGCUCAUCCAUGCUGGAGAGUAGUAAGUUAACGGAA-AAG------UCCUUUAAUUAACACU .(((...((.((((((...(((.((((((....))))))((((.(((.....))))))))))..)))))).)).-..)------)).............. ( -27.90) >DroGri_CAF1 78481 95 + 1 CGGCAUGUCGGUGAAUACGGCACACGCCAAUAAUGGUGUGCUCAUCCAUGCGGGCGAGUAGUAAGUA----CAA-AACACAUACACAUACAAUUUGAACU ..((((((((.......)))).(((((((....))))))).......))))(..(((((.(((.((.----...-.........)).))).)))))..). ( -23.12) >DroEre_CAF1 69798 92 + 1 AGACAUGUCGGUUAAUACGGCUCACGCCAAUAAUGGCGUGCUCAUCCAUGCUGGAGAGUAGUAAGUUGCCGGAA--AC------UCCUUAAAUUGCCACU .((....))((......((((..((((((....))))))((((.(((.....)))))))........))))...--..------.))............. ( -26.00) >DroYak_CAF1 69094 94 + 1 AGACAUGUCGGUUAAUACGGCUCACGCCAACAAUGGCGUGCUCAUCCACGCUGGAGAGUAGUAAGUUGCCGGAAAAAG------UCCUUAAAUUGAAACG .(((...(((((...(((.((((((((((....)))))).....(((.....))))))).)))....))))).....)------)).............. ( -26.80) >DroMoj_CAF1 74796 78 + 1 CAACAUGUCGAUCAAUACGGCACACGCAAAUAAUGGUGUGCUCAUACAUGCAGGAGAGUAGUAAGUA----U------------------AAUUAGAACU .......((.....(((((((((((.((.....)))))))))..(((.(((......)))))).)))----)------------------.....))... ( -15.00) >DroAna_CAF1 60069 82 + 1 CAGCAUGUCGGUUAACACGGCUCACGCCAACAAUGGCGUGCUUAUCCACGCCGGAGAGUAGUAAGUAG----------------UCCGUGAA--AAUGCA ..((((..((((((..((.((((((((((....)))))).....(((.....))))))).))...)))----------------.)))....--.)))). ( -25.80) >consensus AGACAUGUCGGUUAAUACGGCUCACGCCAACAAUGGCGUGCUCAUCCAUGCUGGAGAGUAGUAAGUAG___GAA__A_______UCCUUAAAUUAAAACU ......((((.......)))).(((((((....)))))))(((.(((.....)))))).......................................... (-16.41 = -16.55 + 0.14)

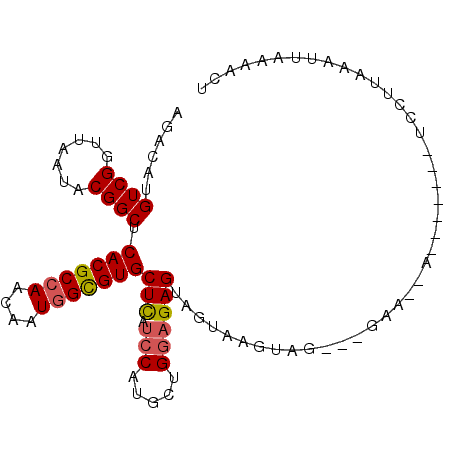
| Location | 12,824,218 – 12,824,311 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -13.58 |
| Energy contribution | -13.63 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
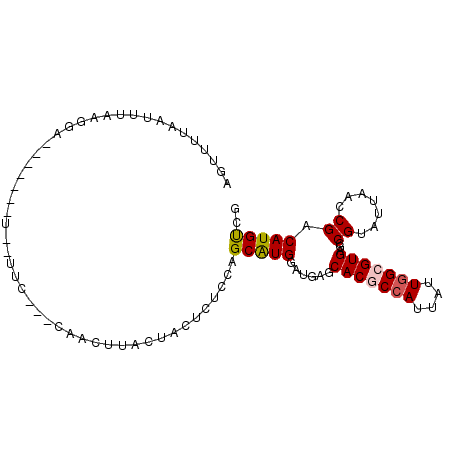
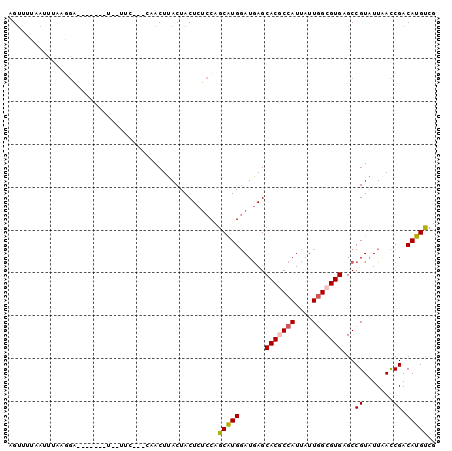
>3L_DroMel_CAF1 12824218 93 - 23771897 AGUGUUAAUUAAAGGA------CUU-UUCCGUUAACUUACUACUCUCCAGCAUGGAUGAGCACGCCAUUGUUGGCGUGAGCCGUAUUAACCGACAUGUCU .(((((.......(((------...-.)))(((((..(((..((((((.....))).)))(((((((....)))))))....)))))))).))))).... ( -26.50) >DroGri_CAF1 78481 95 - 1 AGUUCAAAUUGUAUGUGUAUGUGUU-UUG----UACUUACUACUCGCCCGCAUGGAUGAGCACACCAUUAUUGGCGUGUGCCGUAUUCACCGACAUGCCG .(((((...(.((((((..((.((.-..(----(....)).)).))..)))))).))))))...........(((((((.(.((....)).)))))))). ( -22.80) >DroEre_CAF1 69798 92 - 1 AGUGGCAAUUUAAGGA------GU--UUCCGGCAACUUACUACUCUCCAGCAUGGAUGAGCACGCCAUUAUUGGCGUGAGCCGUAUUAACCGACAUGUCU ...((((......((.------..--...((((.........((((((.....))).)))(((((((....))))))).))))......))....)))). ( -29.10) >DroYak_CAF1 69094 94 - 1 CGUUUCAAUUUAAGGA------CUUUUUCCGGCAACUUACUACUCUCCAGCGUGGAUGAGCACGCCAUUGUUGGCGUGAGCCGUAUUAACCGACAUGUCU (((.((.......(((------.....)))(((.........((((((.....))).)))(((((((....))))))).))).........)).)))... ( -27.60) >DroMoj_CAF1 74796 78 - 1 AGUUCUAAUU------------------A----UACUUACUACUCUCCUGCAUGUAUGAGCACACCAUUAUUUGCGUGUGCCGUAUUGAUCGACAUGUUG ..........------------------.----................(((((((((.((((((((.....)).)))))))))........)))))).. ( -15.50) >DroAna_CAF1 60069 82 - 1 UGCAUU--UUCACGGA----------------CUACUUACUACUCUCCGGCGUGGAUAAGCACGCCAUUGUUGGCGUGAGCCGUGUUAACCGACAUGCUG .((((.--(((((((.----------------((.((((((((.(....).)))).))))(((((((....))))))))))))))......)).)))).. ( -27.60) >consensus AGUUUUAAUUUAAGGA_______U__UUC___CAACUUACUACUCUCCAGCAUGGAUGAGCACGCCAUUAUUGGCGUGAGCCGUAUUAACCGACAUGUCG .................................................(((((......(((((((....)))))))...((.......)).))))).. (-13.58 = -13.63 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:48 2006