| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,818,752 – 12,818,870 |
| Length | 118 |
| Max. P | 0.943892 |

| Location | 12,818,752 – 12,818,870 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -23.58 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12818752 118 + 23771897 AUUUCGCUUAAGUAACUGGCUAUGUUGGCGAUUUCAACGCCAUAGGGAUGCAGCACCAGAUAGGCUGAGGCCAGAGCAAUUAUCUUGGCUUCAGACAAGACUAGCGACUUGAAAGUCA .....(((..(((..((((...((((((((.......))))).......)))...))))...(.(((((((((((.......)).))))))))).)...))))))((((....)))). ( -35.81) >DroVir_CAF1 76451 118 + 1 AUUCCACGCAGAUAGCUGGCAAUGCUGGCAAUGUCGACGCCAUGCGGAUGCAGCACCAGAUAUGCGGAGGUCAACGCUAUUAUCUUGGCUUCAGCUAGCGCCAGCGACAUGAAGUUGC ......(((...(((((((((.(((((((.........)))).)))..)))(((.(.(((((.(((........)))...))))).)))).)))))))))...(((((.....))))) ( -38.20) >DroPse_CAF1 86179 118 + 1 AUUCCGUGGAGGUAGCUGGCAAUGCUGGCUAUCUCCAAGCCAUGGGGAUGCAGCACCAGAUAGGCCGAGGUCAAGGCUAUAACCUUGGCCUCUGCCAGGGCCAGGGAUUUGAAGUUUU (((((.((((((((((..((...))..)))))))))).(((.(((..........)))....(((.((((((((((......)))))))))).)))..)))..))))).......... ( -56.90) >DroSec_CAF1 59961 118 + 1 AUUUCGCUUAAGUAACUGGCUAUGUUGGCGAUGUCAACGCCAUGGGGAUGCAGCACCAGAUAGGCUGAGGCUAGAGCAAUUAUCUUGGCCUCGGUCAGGACUAGCGACUUGAAAGUCA .....(((..(((..((((...((((((((.......))))).......)))...))))...(((((((((((((.......)).)))))))))))...))))))((((....)))). ( -39.91) >DroMoj_CAF1 69149 118 + 1 AUUCCACGUAAAUAGCUGGAAAUGCUGUCAAUGUCCACGCCAUGGGGAUGCAGGACCAAAUAGGCGGAAGUCAAUGCUAUAAUCUUAGCUUCAAAAAGUGCCAGCGACAUUAGCUCAU .(((((.((.....)))))))..((((...(((((..(.(....).)..((.((..(.....(((....)))...((((......))))........)..)).))))))))))).... ( -27.30) >DroPer_CAF1 84477 118 + 1 AUUCCGUGGAGGUAGCUGGCAAUGCUGGCUAUCUCCAAGCCAUGGGGAUGCAGCACCAGAUAGGCCGAGGUCAAGGCUAUAACCUUGGCCUCUGCCAGGGCCAGGGAUUUGAAGUUUU (((((.((((((((((..((...))..)))))))))).(((.(((..........)))....(((.((((((((((......)))))))))).)))..)))..))))).......... ( -56.90) >consensus AUUCCGCGUAAGUAGCUGGCAAUGCUGGCAAUGUCCACGCCAUGGGGAUGCAGCACCAGAUAGGCCGAGGUCAAAGCUAUAAUCUUGGCCUCAGCCAGGGCCAGCGACUUGAAGUUCU .........((((..((((...(.(((((.........(((((..((........))..)).))).((((((((..........)))))))).))))).)))))..))))........ (-23.58 = -22.70 + -0.88)


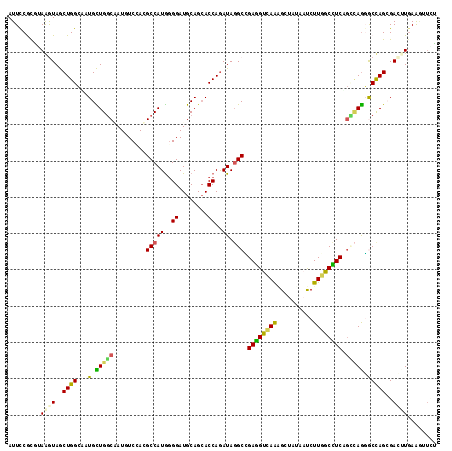
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:45 2006