
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,808,506 – 12,808,603 |
| Length | 97 |
| Max. P | 0.793170 |

| Location | 12,808,506 – 12,808,603 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -16.81 |
| Energy contribution | -18.62 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
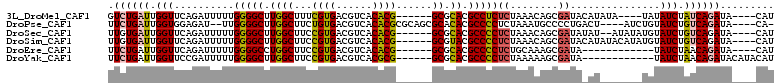
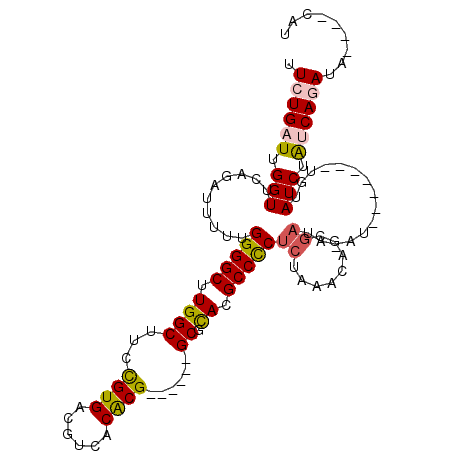
>3L_DroMel_CAF1 12808506 97 + 23771897 AUG----UAUCUGAUAGAUAUA----UAUAUGUAUCGCUGUUUAGAGAGGCGUGCGC------CGUGUGACGUCACGAAAGCCAAGCCCCAAAAAUCUGAACCAAUCAGAC ...----..((((((.((((((----....))))))...(((((((..(((.((.((------..((((....))))...)))).))).......)))))))..)))))). ( -27.20) >DroPse_CAF1 76304 100 + 1 -UG----UAUCUGACAGAUACAGAU----AGUCAGGGGCAUUUAGAGGGGCGUGUGCGCUGCGCGUGUGACGUCACAGAAGCCAAGCCCCAA--AUCUCCACCAAUCAGAA -((----((((.....))))))(((----.(...((((.((((...(((((...((.(((...(.((((....))))).))))).)))))))--))))))..).))).... ( -31.40) >DroSec_CAF1 49966 99 + 1 AUG----UAUCUGACAGAUACAUAUAU--AUAUAUCGCUGUUUAGAGGGGCGUGCGC------CGUGUGACGUCACGGAAGCCAAGCCCCAAAAAUCUGAACCAAUCACAA (((----((((.....)))))))....--..........((((((((((((..((.(------((((......)))))..))...))))).....)))))))......... ( -32.60) >DroSim_CAF1 49383 101 + 1 AUG----UAUCUGACAGAUACAUAUGUAUAUGUAUCGCUGUUUAGAGGGGCGUACGC------CGUGUGACGUCACGGAAGCCAAGCCCCAAAAAUCUGAACCAAUCACAA .((----(........((((((((....))))))))...((((((((((((.....(------((((......))))).......))))).....))))))).....))). ( -33.00) >DroEre_CAF1 54031 89 + 1 AUG----UAUCUGUUAGAUA------------UAUCGCUUUGCAGAGGGGCGUGCGC------CGUGUGACGUCACGGAAGCCAGGCCCCAAAAAUCUGAACCAAUCAGAA ...----..(((((.((...------------.....))..)))))(((((.((..(------((((......)))))....)).))))).....(((((.....))))). ( -31.60) >DroYak_CAF1 53094 93 + 1 AUGUAUGUAUCUGUUAGAUA------------UAUCGCUUUUUAGAGGGGCGUGCGC------CGCGUGACGUCACGGAAGCCAAGCCCCAAAAAUCGGAACCAAUCAGAA ..(((((((((.....))))------------))).))........(((((.((.((------..((((....))))...)))).)))))..................... ( -25.30) >consensus AUG____UAUCUGACAGAUACA_______AU_UAUCGCUGUUUAGAGGGGCGUGCGC______CGUGUGACGUCACGGAAGCCAAGCCCCAAAAAUCUGAACCAAUCAGAA .........(((((..((((............)))).....)))))(((((.((.((......((((......))))...)))).))))).....(((((.....))))). (-16.81 = -18.62 + 1.81)



| Location | 12,808,506 – 12,808,603 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12808506 97 - 23771897 GUCUGAUUGGUUCAGAUUUUUGGGGCUUGGCUUUCGUGACGUCACACG------GCGCACGCCUCUCUAAACAGCGAUACAUAUA----UAUAUCUAUCAGAUA----CAU (((((((..(((.(((.....(((((.(((((...(((....)))..)------)).)).)))))))).)))...((((......----..)))).))))))).----... ( -24.90) >DroPse_CAF1 76304 100 - 1 UUCUGAUUGGUGGAGAU--UUGGGGCUUGGCUUCUGUGACGUCACACGCGCAGCGCACACGCCCCUCUAAAUGCCCCUGACU----AUCUGUAUCUGUCAGAUA----CA- .((((((.((((.((((--..(((((.((((..(((((.((.....))))))).)).)).))))).................----)))).)))).))))))..----..- ( -35.41) >DroSec_CAF1 49966 99 - 1 UUGUGAUUGGUUCAGAUUUUUGGGGCUUGGCUUCCGUGACGUCACACG------GCGCACGCCCCUCUAAACAGCGAUAUAU--AUAUAUGUAUCUGUCAGAUA----CAU .((((.((((((.(((.....(((((...((..(((((......))))------).))..)))))))).)))...(((((((--....)))))))...))).))----)). ( -30.20) >DroSim_CAF1 49383 101 - 1 UUGUGAUUGGUUCAGAUUUUUGGGGCUUGGCUUCCGUGACGUCACACG------GCGUACGCCCCUCUAAACAGCGAUACAUAUACAUAUGUAUCUGUCAGAUA----CAU .((((.((((((.(((.....(((((...((..(((((......))))------).))..)))))))).)))...((((((((....))))))))...))).))----)). ( -31.60) >DroEre_CAF1 54031 89 - 1 UUCUGAUUGGUUCAGAUUUUUGGGGCCUGGCUUCCGUGACGUCACACG------GCGCACGCCCCUCUGCAAAGCGAUA------------UAUCUAACAGAUA----CAU .(((((.....))))).....(((((...((..(((((......))))------).))..)))))..............------------((((.....))))----... ( -26.20) >DroYak_CAF1 53094 93 - 1 UUCUGAUUGGUUCCGAUUUUUGGGGCUUGGCUUCCGUGACGUCACGCG------GCGCACGCCCCUCUAAAAAGCGAUA------------UAUCUAACAGAUACAUACAU .((((.((((...((.((((((((((.(((((..((((....)))).)------)).)).)))))...))))).))...------------...))))))))......... ( -27.10) >consensus UUCUGAUUGGUUCAGAUUUUUGGGGCUUGGCUUCCGUGACGUCACACG______GCGCACGCCCCUCUAAACAGCGAUA_AU_______UGUAUCUAUCAGAUA____CAU .((((((.(((..........(((((.((((...((((......))))......)).)).)))))((........))...............))).))))))......... (-18.10 = -18.30 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:40 2006