
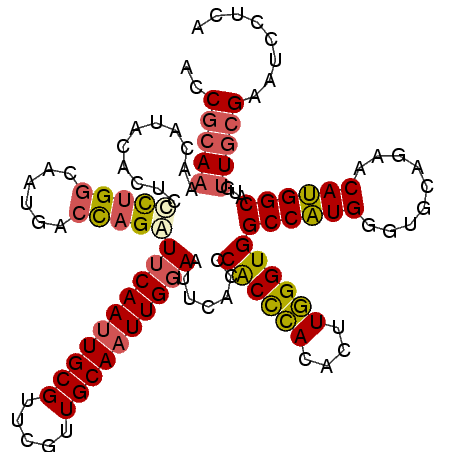
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,798,320 – 12,798,439 |
| Length | 119 |
| Max. P | 0.968470 |

| Location | 12,798,320 – 12,798,439 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.10 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -34.66 |
| Energy contribution | -34.88 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12798320 119 + 23771897 ACCGCAAAACAUACACUCCCUGGCAAUGACCAGGUUCAAUUGCGUUCGUUGCAAUUGGAAUUCACCCACCCACACUUGGGUGGCCAUGGGUGCGGAACAUGGCAUGUUGCGAAUCCUCA ..((((..((((......(((((......)))))((((((((((.....)))))))))).......((((((....))))))((((((.........))))))))))))))........ ( -43.00) >DroSec_CAF1 39716 119 + 1 ACCGCAAAACAUACACUCCCUGGCAUUGACCAGAUUCAAUUGCGUUCGUUGCAAUUGGAAUUCACCCACCCACACUUGGGUGGCCAUGGGUGCAGAACAUGGCAUGUUGCGAAUCCUCA ..((((..((((.......((((......)))).((((((((((.....)))))))))).......((((((....))))))((((((.........))))))))))))))........ ( -40.30) >DroSim_CAF1 39007 119 + 1 ACCGCAAAACAUACACUCCCUGGCAUUGACCAGAUUCAAUUGCGUUCGUUGCAAUUGGAAUUCACCCACCCACACUUGGGUGGCCAUGGGUGCAGAACAUGGCAUGUUGCGAAUCCUCA ..((((..((((.......((((......)))).((((((((((.....)))))))))).......((((((....))))))((((((.........))))))))))))))........ ( -40.30) >DroEre_CAF1 43931 119 + 1 ACCGCAAAACAUAAACUCUCUGGCAAUGACCAGAUUCAAUUGCGUUCGAUGCAAUUGGAAUACGUCCACCCAUACUUGGGUGGCCAUGGGUGCCGAACAUGGCAUGUUGCGAAUCCUCA ..((((..((((..(((((((((......)))))(((((((((((...)))))))))))....(.(((((((....))))))).)..))))(((......)))))))))))........ ( -43.10) >DroYak_CAF1 42220 119 + 1 ACCGCAAAACAUACACUCUCUGGCACUGACCAGAUUCAAUUGCGUUCGUUGCAAUUGGAAUUCCUCCACUCAUACGUGGGUGGCCAUGGGCGCAGAACAUGGCUUGUUGCGAAUCCUUA ..(((((......(((..(((((......)))))((((((((((.....))))))))))......((((......)))))))((((((..(...)..))))))...)))))........ ( -39.40) >DroAna_CAF1 32194 119 + 1 GCCAAAAAACCUUACCAGUUUGCCCCUGACUCGAUACAACUGCGUUCACUGCAAUUGGAACAGUCCCGCCAACUCCUUGGUGGCCGUGGGCACCGAACACGGCCUGCUGCGUAUUCUCA ............(((((((.((((((.((((.....(((.((((.....)))).)))....))))(((((((....)))))))..).)))))(((....)))...)))).)))...... ( -32.70) >consensus ACCGCAAAACAUACACUCCCUGGCAAUGACCAGAUUCAAUUGCGUUCGUUGCAAUUGGAAUUCACCCACCCACACUUGGGUGGCCAUGGGUGCAGAACAUGGCAUGUUGCGAAUCCUCA ..(((((...........(((((......)))))((((((((((.....)))))))))).......((((((....))))))((((((.........))))))...)))))........ (-34.66 = -34.88 + 0.23)

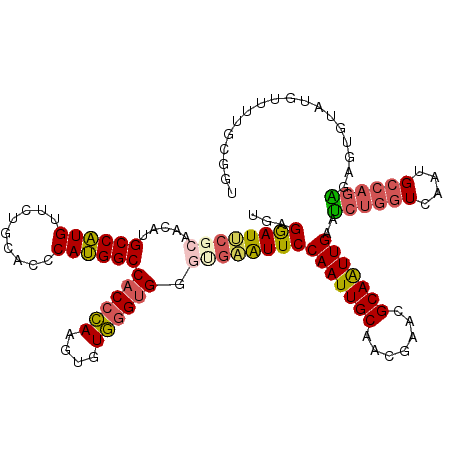
| Location | 12,798,320 – 12,798,439 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.10 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -33.51 |
| Energy contribution | -33.85 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12798320 119 - 23771897 UGAGGAUUCGCAACAUGCCAUGUUCCGCACCCAUGGCCACCCAAGUGUGGGUGGGUGAAUUCCAAUUGCAACGAACGCAAUUGAACCUGGUCAUUGCCAGGGAGUGUAUGUUUUGCGGU .......(((((((((((.........(((((((..((((......))))))))))).(((((((((((.......)))))))..((((((....))))))))))))))))..))))). ( -44.50) >DroSec_CAF1 39716 119 - 1 UGAGGAUUCGCAACAUGCCAUGUUCUGCACCCAUGGCCACCCAAGUGUGGGUGGGUGAAUUCCAAUUGCAACGAACGCAAUUGAAUCUGGUCAAUGCCAGGGAGUGUAUGUUUUGCGGU .......(((((((((((.........(((((((..((((......))))))))))).(((((((((((.......))))))....(((((....))))))))))))))))..))))). ( -42.00) >DroSim_CAF1 39007 119 - 1 UGAGGAUUCGCAACAUGCCAUGUUCUGCACCCAUGGCCACCCAAGUGUGGGUGGGUGAAUUCCAAUUGCAACGAACGCAAUUGAAUCUGGUCAAUGCCAGGGAGUGUAUGUUUUGCGGU .......(((((((((((.........(((((((..((((......))))))))))).(((((((((((.......))))))....(((((....))))))))))))))))..))))). ( -42.00) >DroEre_CAF1 43931 119 - 1 UGAGGAUUCGCAACAUGCCAUGUUCGGCACCCAUGGCCACCCAAGUAUGGGUGGACGUAUUCCAAUUGCAUCGAACGCAAUUGAAUCUGGUCAUUGCCAGAGAGUUUAUGUUUUGCGGU .......((((((...((((((.........))))))((((((....))))))((((((..((((((((.......)))))))..((((((....))))))..)..)))))))))))). ( -41.40) >DroYak_CAF1 42220 119 - 1 UAAGGAUUCGCAACAAGCCAUGUUCUGCGCCCAUGGCCACCCACGUAUGAGUGGAGGAAUUCCAAUUGCAACGAACGCAAUUGAAUCUGGUCAGUGCCAGAGAGUGUAUGUUUUGCGGU .......((((((..(..(((.(((((.((...((((((.((((......))))........(((((((.......)))))))....))))))..))))))).)))..)...)))))). ( -37.60) >DroAna_CAF1 32194 119 - 1 UGAGAAUACGCAGCAGGCCGUGUUCGGUGCCCACGGCCACCAAGGAGUUGGCGGGACUGUUCCAAUUGCAGUGAACGCAGUUGUAUCGAGUCAGGGGCAAACUGGUAAGGUUUUUUGGC ..........(((.(((((....(((((((((.(.((((.(.....).)))).)(((((...(((((((.(....))))))))...).))))..))))..)))))...))))).))).. ( -37.90) >consensus UGAGGAUUCGCAACAUGCCAUGUUCUGCACCCAUGGCCACCCAAGUGUGGGUGGGUGAAUUCCAAUUGCAACGAACGCAAUUGAAUCUGGUCAAUGCCAGAGAGUGUAUGUUUUGCGGU ...((((((((.....((((((.........))))))((((((....)))))).))))))))(((((((.......)))))))..((((((....)))))).................. (-33.51 = -33.85 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:34 2006