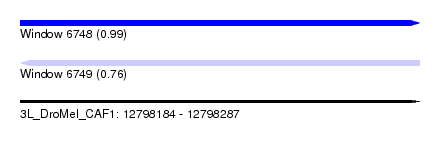
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,798,184 – 12,798,287 |
| Length | 103 |
| Max. P | 0.985732 |

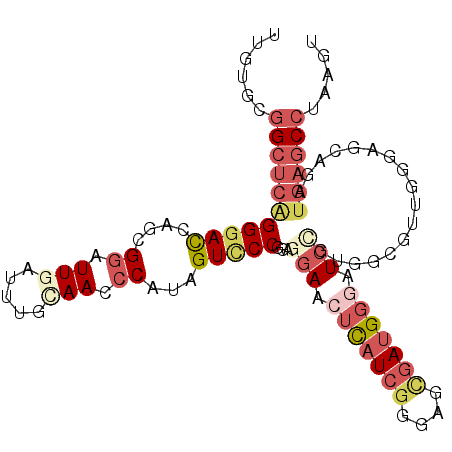
| Location | 12,798,184 – 12,798,287 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -24.72 |
| Energy contribution | -25.37 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.44 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12798184 103 + 23771897 ACUUAGGCUCAUCUGCCCCCAGCGCAACGGAUCCCAUCGAUCCCGAUGAGUUCGUCCGGGACUAUGGGUUGCAAAUCAAUCCGCUGGUCCCUGAGCCGCACAA .....((((((..(((.......)))((((((..(((((....))))).))))))..((((((((((((((.....))))))).)))))))))))))...... ( -40.30) >DroSec_CAF1 39580 103 + 1 AUUUAGGCUCAUCUGCUCCCAACGCAACGGAUCCCAUCGGUCCCGAUGAGUUCCUACGGGACUAUGGGUUACAAAUCAAUCCGCUGGUCCCUGAGCCGCACAA .....((((((..(((.......)))..(((.(.(((((....))))).).)))...(((((((((((((.......)))))).)))))))))))))...... ( -39.30) >DroSim_CAF1 38871 103 + 1 ACUUAGGCUCAUCUGCUCCCAACGCAACAGAUCCCAUCGGUCCCGAUGAGUUCCUACGGGACUAUGGAUUGCAAAUCAAUCCGCUGGUCCCUGAGCCGCACAA .....((((((..(((.......)))...((.(.(((((....))))).).))....((((((((((((((.....))))))).)))))))))))))...... ( -38.70) >DroEre_CAF1 43795 103 + 1 AUUUAGGCUCGUCUGCUCCGAAGACCACGGAUCCCAUCACCCCCGAUGAGUUCCUCCGGAACUACGGGUUGCAAAUCAAUCCCCUGAUCCCUGAGCCCCACAA .....(((((((((.......))))...(((((.((((......))))((((((...))))))..((((((.....))))))...)))))..)))))...... ( -32.80) >DroYak_CAF1 42084 103 + 1 ACCUGGGCUCAUCUGCUCCGAAGGCCACGGAUCCCAUCACCCCCGAUGACUUCCUCCGGGACUAUGGGUUGCAAAUUAAUCCACUGGUCCCUGAGCCCCACAA ....(((((((...(((.....)))...(((...((((......))))...)))...((((((((((((((.....))))))).))))))))))))))..... ( -38.30) >DroAna_CAF1 32058 103 + 1 ACUUGAGUGAAGCAACUUCAGAUCCCAAGGAUCCCAUUACCCCGGAGACCUUUUUUCGGGACUAUGCUUUGCGAAUAAAGCCUGUAGUUCCCGUGCAACCGAA ......(..(((((......(((((...))))).......((((((((....))))))))....)))))..).......((.((.......)).))....... ( -25.20) >consensus ACUUAGGCUCAUCUGCUCCCAACGCAACGGAUCCCAUCACCCCCGAUGAGUUCCUCCGGGACUAUGGGUUGCAAAUCAAUCCGCUGGUCCCUGAGCCGCACAA .....((((((.................(((...((((......))))...)))...((((((((((((((.....))))))).)))))))))))))...... (-24.72 = -25.37 + 0.65)

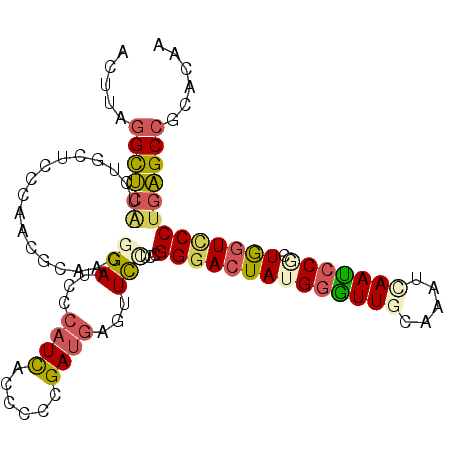
| Location | 12,798,184 – 12,798,287 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -21.01 |
| Energy contribution | -23.18 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12798184 103 - 23771897 UUGUGCGGCUCAGGGACCAGCGGAUUGAUUUGCAACCCAUAGUCCCGGACGAACUCAUCGGGAUCGAUGGGAUCCGUUGCGCUGGGGGCAGAUGAGCCUAAGU ......((((((..(.(((((((.(((.....))).))...((..((((....(((((((....))))))).))))..)))))))...)...))))))..... ( -39.80) >DroSec_CAF1 39580 103 - 1 UUGUGCGGCUCAGGGACCAGCGGAUUGAUUUGUAACCCAUAGUCCCGUAGGAACUCAUCGGGACCGAUGGGAUCCGUUGCGUUGGGAGCAGAUGAGCCUAAAU ......((((((..(((((((((((..........(((((.((((((...(....)..))))))..))))))))))))).)))(....)...))))))..... ( -39.40) >DroSim_CAF1 38871 103 - 1 UUGUGCGGCUCAGGGACCAGCGGAUUGAUUUGCAAUCCAUAGUCCCGUAGGAACUCAUCGGGACCGAUGGGAUCUGUUGCGUUGGGAGCAGAUGAGCCUAAGU ......(((((.(((((.(..((((((.....)))))).).))))).......(((((((....)))))))(((((((.(....).))))))))))))..... ( -44.40) >DroEre_CAF1 43795 103 - 1 UUGUGGGGCUCAGGGAUCAGGGGAUUGAUUUGCAACCCGUAGUUCCGGAGGAACUCAUCGGGGGUGAUGGGAUCCGUGGUCUUCGGAGCAGACGAGCCUAAAU .....((((((.(((((.(.(((.(((.....)))))).).)))))...(((.(((((((....))))))).))).((.((....)).))...)))))).... ( -40.30) >DroYak_CAF1 42084 103 - 1 UUGUGGGGCUCAGGGACCAGUGGAUUAAUUUGCAACCCAUAGUCCCGGAGGAAGUCAUCGGGGGUGAUGGGAUCCGUGGCCUUCGGAGCAGAUGAGCCCAGGU .....((((((((((((..((((.............)))).))))).......((..((((((((.((((...)))).)))))))).))...))))))).... ( -41.42) >DroAna_CAF1 32058 103 - 1 UUCGGUUGCACGGGAACUACAGGCUUUAUUCGCAAAGCAUAGUCCCGAAAAAAGGUCUCCGGGGUAAUGGGAUCCUUGGGAUCUGAAGUUGCUUCACUCAAGU ...(((.((.(((((.(((...(((((......))))).)))))))).....(((((((.(((((......))))).)))))))...)).))).......... ( -31.20) >consensus UUGUGCGGCUCAGGGACCAGCGGAUUGAUUUGCAACCCAUAGUCCCGGAGGAACUCAUCGGGAGCGAUGGGAUCCGUGGCGUUGGGAGCAGAUGAGCCUAAGU ......(((((((((((....((.(((.....))).))...)))))...(((.(((((((....))))))).))).................))))))..... (-21.01 = -23.18 + 2.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:32 2006