
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,787,775 – 12,787,882 |
| Length | 107 |
| Max. P | 0.981360 |

| Location | 12,787,775 – 12,787,882 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -29.12 |
| Energy contribution | -29.07 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
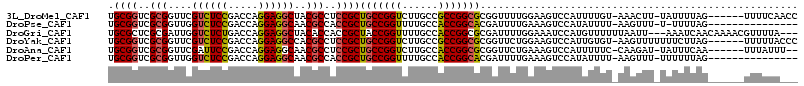
>3L_DroMel_CAF1 12787775 107 + 23771897 UGCGGUCGCGGUUCGUCUCCGACCAGGAGGCUACGCCUCCGCUGCCGGUCUUGCCGCCGGCGCGGUUUUGGAAGUCCAUUUUGU-AAACUU-UAUUUUAG------UUUUCAACC .((((..(((....((((((.....))))))..)))..)))).((((((......))))))..((((.(((....)))....(.-(((((.-......))------))).))))) ( -39.80) >DroPse_CAF1 54304 97 + 1 UGCGGUCGCGGUUGGUCUCCGACCAGGAGGCAACGCCACCGCUGCCGGUUUUGCCACCGGCACGAUUUUGAAAGUCCAUAUUUU-AAGUUU-U-UUUUAG--------------- .(((((.(((....((((((.....))))))..))).)))))(((((((......)))))))....(((((((.......))))-)))...-.-......--------------- ( -37.40) >DroGri_CAF1 44192 109 + 1 UGCGCUCGCGAUUGGUCUCUGACCAGGAGGCUACACCACCGCUACCGGUUUUGCCACCGGCGCGAUUUUGGAAAUCCAUGUUUUUUAAUU---AAAUCAACAAAACGUUUUA--- .(((((......(((((...)))))((.(((......((((....))))...))).)))))))(((((..((((........))))....---)))))..............--- ( -27.00) >DroYak_CAF1 31687 108 + 1 UGCGGUCGCGGUUCGUCUCCGACCAGGAGGCCACGCCUCCGCUGCCGGUCUUGCCGCCGGCGCGGUUCUGGAAGUCCAUUGUGU-AAGUUUUUUUCUUAG------UUUUUACCC .((((..(((....((((((.....))))))..)))..)))).((((((......))))))..(((..(((....))).....(-(((.......)))).------.....))). ( -38.30) >DroAna_CAF1 22138 105 + 1 UGCGGUCGCGGUUCGAUUCCGACCAGGAGGCAACGCCUCCGCUGCCGGUCUUGCCACCGGCGCGGUUCUGAAAGUCCAUUUUUC-CAAGAU-UAUUUCAA------UUUAUUU-- ...(((((.((......))))))).((((((...))))))((.((((((......))))))))...((((((((.....)))))-..))).-........------.......-- ( -34.80) >DroPer_CAF1 52624 98 + 1 UGCGGUCGCGGUUGGUCUCCGACCAGGAGGCAACGCCACCGCUGCCGGUUUUGCCACCGGCACGAUUUUGAAAGUCCAUAUUUU-AAGUUU-UUUUUUAG--------------- .(((((.(((....((((((.....))))))..))).)))))(((((((......)))))))....(((((((.......))))-)))...-........--------------- ( -37.40) >consensus UGCGGUCGCGGUUCGUCUCCGACCAGGAGGCAACGCCACCGCUGCCGGUCUUGCCACCGGCGCGAUUUUGAAAGUCCAUUUUUU_AAAUUU_UAUUUUAG______UUUUUA___ .((((..(((....((((((.....))))))..)))..))))(((((((......)))))))..................................................... (-29.12 = -29.07 + -0.05)


| Location | 12,787,775 – 12,787,882 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -26.33 |
| Energy contribution | -26.25 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12787775 107 - 23771897 GGUUGAAAA------CUAAAAUA-AAGUUU-ACAAAAUGGACUUCCAAAACCGCGCCGGCGGCAAGACCGGCAGCGGAGGCGUAGCCUCCUGGUCGGAGACGAACCGCGACCGCA (((((.(((------((......-.)))))-.)....(((....))).))))((((((((.....).))))).((((((((...)))))).(((((....)).)))))....)). ( -42.20) >DroPse_CAF1 54304 97 - 1 ---------------CUAAAA-A-AAACUU-AAAAUAUGGACUUUCAAAAUCGUGCCGGUGGCAAAACCGGCAGCGGUGGCGUUGCCUCCUGGUCGGAGACCAACCGCGACCGCA ---------------......-.-......-......................(((((((......)))))))(((((.((((((.((((.....))))..))).))).))))). ( -36.00) >DroGri_CAF1 44192 109 - 1 ---UAAAACGUUUUGUUGAUUU---AAUUAAAAAACAUGGAUUUCCAAAAUCGCGCCGGUGGCAAAACCGGUAGCGGUGGUGUAGCCUCCUGGUCAGAGACCAAUCGCGAGCGCA ---......((((((..(((((---(...........))))))..))))))(((((((((......)))))).((((((((.(.(((....)))...).))).)))))..))).. ( -32.80) >DroYak_CAF1 31687 108 - 1 GGGUAAAAA------CUAAGAAAAAAACUU-ACACAAUGGACUUCCAGAACCGCGCCGGCGGCAAGACCGGCAGCGGAGGCGUGGCCUCCUGGUCGGAGACGAACCGCGACCGCA .(((.....------.((((.......)))-).....(((....)))..)))((((((((.....).))))).((((((((...)))))).(((((....)).)))))....)). ( -39.60) >DroAna_CAF1 22138 105 - 1 --AAAUAAA------UUGAAAUA-AUCUUG-GAAAAAUGGACUUUCAGAACCGCGCCGGUGGCAAGACCGGCAGCGGAGGCGUUGCCUCCUGGUCGGAAUCGAACCGCGACCGCA --.......------((((((..-.(((..-.......))).))))))......((((((......)))))).((((((((...)))))).((((((........).))))))). ( -34.60) >DroPer_CAF1 52624 98 - 1 ---------------CUAAAAAA-AAACUU-AAAAUAUGGACUUUCAAAAUCGUGCCGGUGGCAAAACCGGCAGCGGUGGCGUUGCCUCCUGGUCGGAGACCAACCGCGACCGCA ---------------........-......-......................(((((((......)))))))(((((.((((((.((((.....))))..))).))).))))). ( -36.00) >consensus ___UAAAAA______CUAAAAUA_AAACUU_AAAAAAUGGACUUCCAAAACCGCGCCGGUGGCAAAACCGGCAGCGGAGGCGUUGCCUCCUGGUCGGAGACCAACCGCGACCGCA ......................................(((.........((((((((((......)))))).)))).(((...)))))).((((((........).)))))... (-26.33 = -26.25 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:27 2006