
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,778,253 – 12,778,388 |
| Length | 135 |
| Max. P | 0.979283 |

| Location | 12,778,253 – 12,778,348 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -16.73 |
| Energy contribution | -16.12 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12778253 95 + 23771897 UAGGUAAAUGGAAGGUAUUUAUUUGUUUUGAAGAGUUUAUUUUCUUACCUUGCGAUGACGUCCUUUGUAGACGACGGCAAAGGCGCCAUGGCCCA .(((((...((((((((...((((........)))).))))))))))))).((.(((.((.(((((((........))))))))).))).))... ( -26.30) >DroVir_CAF1 39895 82 + 1 UAGAUAAAC--AAGAUAUUG---------AUGUGA--CGUUUGCUUACCUUGCGAUGACGUCCUUUGUAGACCACAGCAAAGGCGCCAUGGCCAA ........(--((....)))---------..((((--.(....)))))...((.(((.((.(((((((........))))))))).))).))... ( -17.00) >DroPse_CAF1 45844 88 + 1 -----AAAU-GACGAAAUUAAAGUGUUUUUU-UAUUAUUAGUUCUUACCUUGCGGUGACGCCCUUUGUAGACGACAGCAAAGGCGCCAUGGCCAA -----.(((-((.(((((......))))).)-))))...............((.(((.((((.(((((........))))))))).))).))... ( -20.00) >DroSec_CAF1 20206 95 + 1 UAGGUAAAUGGAAGAUAUUUAUUUGUUUUGAAGAGUUUCGCUUCUUACCUUGCGAUGACGUCCUUUGUAGACGACGGCAAAGGCGCCGUGGCCCA .((((((....((((((......))))))((((.......)))))))))).((.(((.((.(((((((........))))))))).))).))... ( -25.30) >DroSim_CAF1 19530 95 + 1 UAGGUAAAUAGAAGAUAUUUAUUUGUUUUGAAGAGUUUCGCUUCUUACCUUGCGAUGACGUCCUUUGUAGACGACGGCAAAGGCGCCGUGGCCCA .((((((....((((((......))))))((((.......)))))))))).((.(((.((.(((((((........))))))))).))).))... ( -25.30) >DroMoj_CAF1 33287 90 + 1 CAGUUAAAC--AAUUAAUUA-GUUGCAUUACAUGG--CAAUUUCUUACCUUGCGGUGACGUCCUUUGUAGACCACGGCGAAGGCGCCGUGGCCAA .......((--((.(((..(-(((((........)--)))))..)))....(((....)))...)))).(.((((((((....)))))))).).. ( -27.20) >consensus UAGGUAAAU_GAAGAUAUUUAUUUGUUUUGAAGAGUUUAAUUUCUUACCUUGCGAUGACGUCCUUUGUAGACGACGGCAAAGGCGCCAUGGCCAA ...................................................((.(((.((.(((((((........))))))))).))).))... (-16.73 = -16.12 + -0.61)


| Location | 12,778,253 – 12,778,348 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -23.74 |
| Consensus MFE | -16.19 |
| Energy contribution | -15.22 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12778253 95 - 23771897 UGGGCCAUGGCGCCUUUGCCGUCGUCUACAAAGGACGUCAUCGCAAGGUAAGAAAAUAAACUCUUCAAAACAAAUAAAUACCUUCCAUUUACCUA (((((.((((((((((((..(....)..)))))).)))))).))(((((((((........)))..............)))))))))........ ( -22.33) >DroVir_CAF1 39895 82 - 1 UUGGCCAUGGCGCCUUUGCUGUGGUCUACAAAGGACGUCAUCGCAAGGUAAGCAAACG--UCACAU---------CAAUAUCUU--GUUUAUCUA ...((.((((((((((((..(....)..)))))).)))))).)).((((((((((...--......---------.......))--)))))))). ( -25.39) >DroPse_CAF1 45844 88 - 1 UUGGCCAUGGCGCCUUUGCUGUCGUCUACAAAGGGCGUCACCGCAAGGUAAGAACUAAUAAUA-AAAAAACACUUUAAUUUCGUC-AUUU----- .((((...(((((((((..(((.....))))))))))))(((....)))..............-..................)))-)...----- ( -21.80) >DroSec_CAF1 20206 95 - 1 UGGGCCACGGCGCCUUUGCCGUCGUCUACAAAGGACGUCAUCGCAAGGUAAGAAGCGAAACUCUUCAAAACAAAUAAAUAUCUUCCAUUUACCUA (((((.((((((....)))))).)))))....(((....(((....)))..((((.......)))).................)))......... ( -21.40) >DroSim_CAF1 19530 95 - 1 UGGGCCACGGCGCCUUUGCCGUCGUCUACAAAGGACGUCAUCGCAAGGUAAGAAGCGAAACUCUUCAAAACAAAUAAAUAUCUUCUAUUUACCUA (((((.((((((....)))))).)))))...(((.....(((....)))..((((.......))))........((((((.....))))))))). ( -21.20) >DroMoj_CAF1 33287 90 - 1 UUGGCCACGGCGCCUUCGCCGUGGUCUACAAAGGACGUCACCGCAAGGUAAGAAAUUG--CCAUGUAAUGCAAC-UAAUUAAUU--GUUUAACUG ..((((((((((....)))))))))).((((.((......))(((.(((((....)))--)).)))........-.......))--))....... ( -30.30) >consensus UGGGCCACGGCGCCUUUGCCGUCGUCUACAAAGGACGUCAUCGCAAGGUAAGAAAAGAAACUAUUCAAAACAAAUAAAUAUCUUC_AUUUACCUA ..(((.((((((....)))))).((((.....)))))))(((....))).............................................. (-16.19 = -15.22 + -0.97)

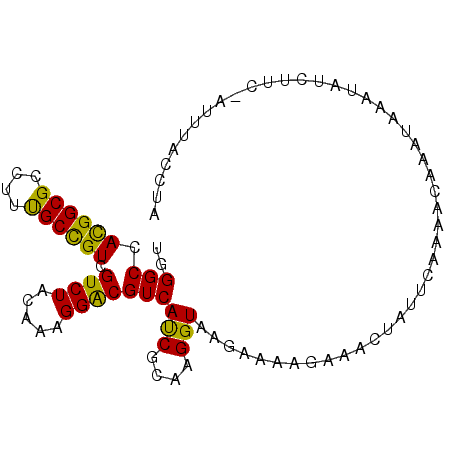

| Location | 12,778,272 – 12,778,388 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -24.00 |
| Energy contribution | -25.03 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12778272 116 - 23771897 AAUAUUGUCGGGGAAUAUGAAUACAGCUCCAAGGAUAUGCUGGGCCAUGGCGCCUUUGCCGUCGUCUACAAAGGACGUCAUCGCAAGGUAAGAAAAUAAACUCUUCAAAACAAAUA ..((((.((((((....((....)).)))).......((((..((.((((((((((((..(....)..)))))).)))))).))..)))).)).)))).................. ( -28.30) >DroVir_CAF1 39912 105 - 1 AAUAUUGUCGGGGAAUAUGAAUACAGCUCCAAGGAUAUGCUUGGCCAUGGCGCCUUUGCUGUGGUCUACAAAGGACGUCAUCGCAAGGUAAGCAAACG--UCACAU---------C .(((((.(.((((....((....)).)))).).)))))(((((((.((((((((((((..(....)..)))))).)))))).))....))))).....--......---------. ( -28.70) >DroPse_CAF1 45857 115 - 1 AAUAUUGUCGGGGAGUAUGAAUACAGCUCCAAGGAUAUGCUUGGCCAUGGCGCCUUUGCUGUCGUCUACAAAGGGCGUCACCGCAAGGUAAGAACUAAUAAUA-AAAAAACACUUU ..((((((..((((((.((....))))))).......(((((.((..((((((((((..(((.....)))))))))))))..)).)))))....)..))))))-............ ( -31.80) >DroWil_CAF1 33381 108 - 1 AAUAUUGUUGGGGACUAUGAAUACAGCUCCAAGGAUAUGCUUGGCCAUGGGGCCUUUGCUGUCGUCUACAAAGGACGUCAUCGCAAGGUAAGU--------UCUUAAUUAUAAUUA ..(((.((((((((((.........((((((.((.((....)).)).))))))((((((((.(((((.....))))).))..))))))..)))--------))))))).))).... ( -34.70) >DroMoj_CAF1 33304 113 - 1 AAUAUUGUCGGGGAAUAUGAAUACAGCUCCAAGGAUAUGCUUGGCCACGGCGCCUUCGCCGUGGUCUACAAAGGACGUCACCGCAAGGUAAGAAAUUG--CCAUGUAAUGCAAC-U .(((((.(.((((....((....)).)))).).)))))((..((((((((((....))))))))))((((..((......))....(((((....)))--)).))))..))...-. ( -38.20) >DroAna_CAF1 14979 108 - 1 AAUAUUGUCGGGGACUAUGAAUACAGCUCCAAGGAUAUGCUUGGCCACGGCGCCUUUGCCGUCGUCUACAAGGGUCGUCAUCGCAAGGUAAGCAUCCAGUCUCCUCUC-------- .........((((((((((..(((.((.(((((......)))))....((((((((((..(....)..)))))).))))...))...)))..)))..)))))))....-------- ( -32.60) >consensus AAUAUUGUCGGGGAAUAUGAAUACAGCUCCAAGGAUAUGCUUGGCCAUGGCGCCUUUGCCGUCGUCUACAAAGGACGUCAUCGCAAGGUAAGAAAACA__CUAUUAAA_ACAA__A .(((((.(.((((....((....)).)))).).)))))((((.((.((((((((((((..(....)..)))))).)))))).)).))))........................... (-24.00 = -25.03 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:23 2006