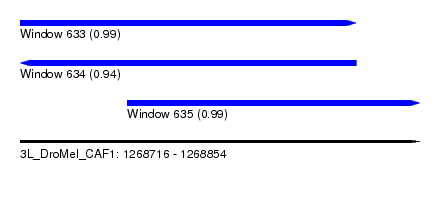
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,268,716 – 1,268,854 |
| Length | 138 |
| Max. P | 0.994090 |

| Location | 1,268,716 – 1,268,832 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.29 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -33.58 |
| Energy contribution | -33.82 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
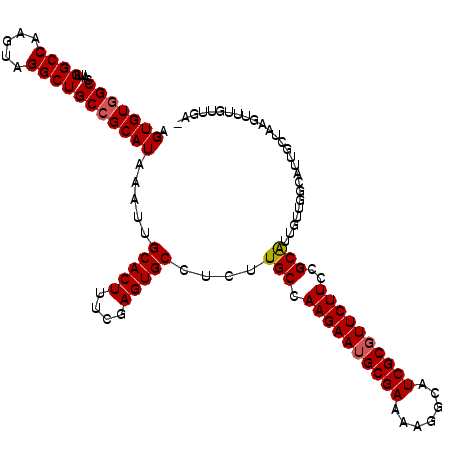
>3L_DroMel_CAF1 1268716 116 + 23771897 AGUGUGGCCAUGGGCCAAGUAGGCUGCCGCAUAAAUUGCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGGCAUCGCGUUCUUCCGCAAUGUUGGCAUUGCUAAGUUUGUUGC- .(((((((....((((.....))))))))))).......((((...((((((.(((((.((((((((((.......))))))))))..)))).)..))))))...)))).......- ( -40.30) >DroSec_CAF1 23487 107 + 1 AGUGUAGCCAUUGGCCAAGUAGGCUGCCGCAUAAAUUGCACUUUCGAGUGCAUCUUGCCAAGAAUGCGAAAAGGAAUCGCGUUCUUCCGCAUU---------GCUAAGUUUGUUUA- .(.(((((((((.....))).)))))))(((.....((((((....))))))...(((.((((((((((.......))))))))))..))).)---------))............- ( -33.80) >DroSim_CAF1 23906 116 + 1 AGUGUGGCCAUUGGCCAAGUAGGCUGCCGCAUAAAUUGCACUUUCGAGUGCAUCUUGCCAAGAAUGCGAAAAGGAAUCGCCUUCUUCCGCAUUGUUGGCAUUGAUAAGUUUGUUUA- .(((((((....((((.....)))))))))))((((((((((....)))))(((.((((((.((((((((((((.....)))).)).)))))).))))))..))).))))).....- ( -44.40) >DroEre_CAF1 23316 117 + 1 AGUGUGGCCAUUGGCCAAGUAGGCUGCCGCAUACAUUGCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGGCAUCGCGUUCUUCCGCAUUGUGGGCAUUGCAGAGGUGGCUGCA ....(((((...))))).....((.((((((.....)))((((..((((((((..(((.((((((((((.......))))))))))..)))....))))))).)..))))))).)). ( -45.90) >DroYak_CAF1 23908 108 + 1 AGUGUGGCCAUUGGCCAAGUAGGCUGCCGCAUAAAUUGCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAGAAGGCAUCGCGUUCUUCCGCGUUGUUGGCAUUGCAGAG--------- .(((((((....((((.....))))))))))).....(((((....)))))((((((((((.((((((.(((((........))))))))))).))))))....))))--------- ( -45.70) >consensus AGUGUGGCCAUUGGCCAAGUAGGCUGCCGCAUAAAUUGCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGGCAUCGCGUUCUUCCGCAUUGUUGGCAUUGCUAAGUUUGUUGA_ .(((((((....((((.....))))))))))).....(((((....)))))....(((.((((((((((.......))))))))))..))).......................... (-33.58 = -33.82 + 0.24)

| Location | 1,268,716 – 1,268,832 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.29 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -28.06 |
| Energy contribution | -29.94 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1268716 116 - 23771897 -GCAACAAACUUAGCAAUGCCAACAUUGCGGAAGAACGCGAUGCCUUUUCGCAUUCUUGGCAAGAGGCACUCGAAAGUGCAAUUUAUGCGGCAGCCUACUUGGCCCAUGGCCACACU -(((.....(((.((((((....))))))..))).((.(((((((((((.((.......)))))))))).)))...))........)))(((.(((.....))).....)))..... ( -35.80) >DroSec_CAF1 23487 107 - 1 -UAAACAAACUUAGC---------AAUGCGGAAGAACGCGAUUCCUUUUCGCAUUCUUGGCAAGAUGCACUCGAAAGUGCAAUUUAUGCGGCAGCCUACUUGGCCAAUGGCUACACU -..........((((---------..(((..(((((.((((.......)))).))))).((((((((((((....))))).)))).))).)))(((.....))).....)))).... ( -30.60) >DroSim_CAF1 23906 116 - 1 -UAAACAAACUUAUCAAUGCCAACAAUGCGGAAGAAGGCGAUUCCUUUUCGCAUUCUUGGCAAGAUGCACUCGAAAGUGCAAUUUAUGCGGCAGCCUACUUGGCCAAUGGCCACACU -................((((((.(((((((((..(((.....)))))))))))).))))))...((((((....)))))).....((.(((.(((.....))).....))).)).. ( -37.60) >DroEre_CAF1 23316 117 - 1 UGCAGCCACCUCUGCAAUGCCCACAAUGCGGAAGAACGCGAUGCCUUUUCGCAUUCUUGGCAAGAGGCACUCGAAAGUGCAAUGUAUGCGGCAGCCUACUUGGCCAAUGGCCACACU .(((((((..((((((.((....)).))))))((((.((((.......)))).)))))))).....(((((....)))))......)))(((.(((.....))).....)))..... ( -39.80) >DroYak_CAF1 23908 108 - 1 ---------CUCUGCAAUGCCAACAACGCGGAAGAACGCGAUGCCUUCUCGCAUUCUUGGCAAGAGGCACUCGAAAGUGCAAUUUAUGCGGCAGCCUACUUGGCCAAUGGCCACACU ---------(((((((.((((((...((((......))))((((......))))..))))))....(((((....)))))......))))).))......(((((...))))).... ( -35.60) >consensus _GAAACAAACUUAGCAAUGCCAACAAUGCGGAAGAACGCGAUGCCUUUUCGCAUUCUUGGCAAGAGGCACUCGAAAGUGCAAUUUAUGCGGCAGCCUACUUGGCCAAUGGCCACACU .............(((.((((((.((((((((((..........)))).)))))).))))))....(((((....)))))......)))(((.(((.....))).....)))..... (-28.06 = -29.94 + 1.88)

| Location | 1,268,753 – 1,268,854 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.71 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -14.42 |
| Energy contribution | -16.62 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
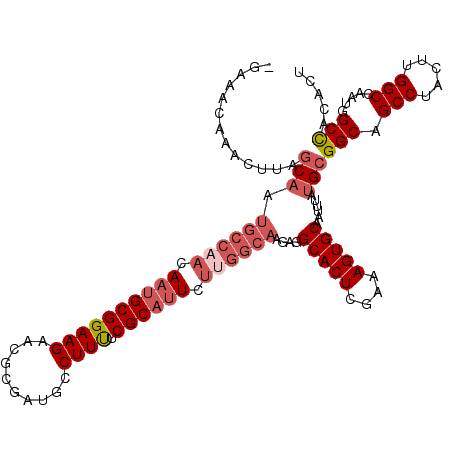
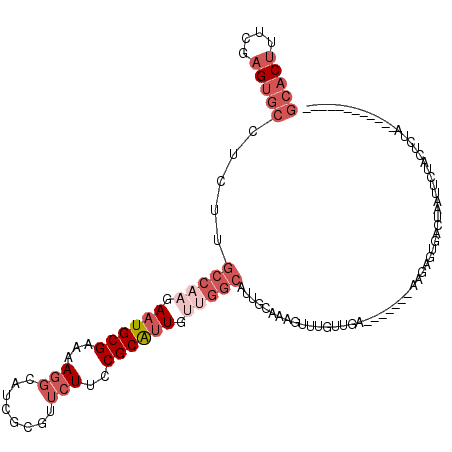
>3L_DroMel_CAF1 1268753 101 + 23771897 GCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGGCAUCGCGUUCUUCCGCAAUGUUGGCAUUGCUAAGUUUGUUGC-------AAGAGUGACUAAUUCUACUCUA------------ (((((....)))))(((((((.((((((((((.......))))))))))..((((((....))))))..........))-------))))).................------------ ( -34.90) >DroSec_CAF1 23524 92 + 1 GCACUUUCGAGUGCAUCUUGCCAAGAAUGCGAAAAGGAAUCGCGUUCUUCCGCAUU---------GCUAAGUUUGUUUA-------AAGGGUGACUAAUUCUACUCUA------------ (((((....)))))....(((.((((((((((.......))))))))))..)))..---------..............-------.((((((........)))))).------------ ( -24.90) >DroSim_CAF1 23943 101 + 1 GCACUUUCGAGUGCAUCUUGCCAAGAAUGCGAAAAGGAAUCGCCUUCUUCCGCAUUGUUGGCAUUGAUAAGUUUGUUUA-------AAGGGUGACUAAUUCUACUCUA------------ (((((....)))))(((.((((((.((((((((((((.....)))).)).)))))).))))))..)))...........-------.((((((........)))))).------------ ( -33.40) >DroEre_CAF1 23353 108 + 1 GCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGGCAUCGCGUUCUUCCGCAUUGUGGGCAUUGCAGAGGUGGCUGCAAUUCGCAGAAACUACUACUUCAACUCUA------------ ........((((((((..(((.((((((((((.......))))))))))..)))....))))......(((((((((((.....))))......))))))).))))..------------ ( -36.20) >DroYak_CAF1 23945 98 + 1 GCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAGAAGGCAUCGCGUUCUUCCGCGUUGUUGGCAUUGCAGAG----------UUUGCGAACAUUACCAAUUCAACUCAA------------ (((((....)))))....((((((.((((((.(((((........))))))))))).))))))((((((..----------.))))))....................------------ ( -31.70) >DroAna_CAF1 24318 101 + 1 GCCCGGCCGAGU-------GCCAAGAAUGCGAAAAUGCAACA----CUGCCGCAUUGCUGGCAUCUCAGAGUCGGUUGU-------AGG-CCAAAACAUGCGAAUCUACGAAAACAAAUU ..((((((((((-------((((..((((((.....(((...----.)))))))))..))))).))).).)))))((((-------(((-(........))....))))))......... ( -30.60) >consensus GCACUUUCGAGUGCCUCUUGCCAAGAAUGCGAAAAGGCAUCGCGUUCUUCCGCAUUGUUGGCAUUGCAAAGUUUGUUGA_______AAGAGUGACUAAUUCUACUCUA____________ (((((....))))).....(((((.((((((...(((........)))..)))))).))))).......................................................... (-14.42 = -16.62 + 2.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:45 2006