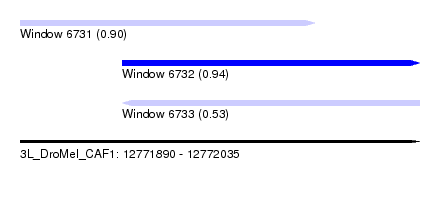
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,771,890 – 12,772,035 |
| Length | 145 |
| Max. P | 0.936130 |

| Location | 12,771,890 – 12,771,997 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 88.50 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -19.02 |
| Energy contribution | -18.98 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12771890 107 + 23771897 GGAAUGACUCAAUUUAAUCGCACGGCAAUUUUAUUGACACAAAACUGACAAAACCAAAUGCGAAAAGGAUUAACAUUUCAAAUUUCGCAUUUGUUUUGCAGGCCAGC ((..((..(((((..(((.((...)).)))..)))))..))...(((.((((((.((((((((((..((........))...))))))))))))))))))).))... ( -27.90) >DroSec_CAF1 13858 107 + 1 GGAAUGACUCAAUUGAAUCGCAUGGGAAUUUAAUUGACACAAAACUGACAAAACCAAAUGCGAAAAGGAUUAACAUUUCCCAUUUCGUAUUUGUUUUGCAGUCCAGC (((.((..(((((((((((......).))))))))))..))...(((.((((((.((((((((((.(((........)))..))))))))))))))))))))))... ( -31.60) >DroSim_CAF1 13228 107 + 1 GGAAUGACUCAAUUGAAUCGCAUGGGAAUUUAAUUGACACAAAACUGACAAAAUCAAAUGCGAAAAGGAUUAACAUUUCCCAUUUCGCAUUUGCUUUGCAGUCCAGC (((.((..(((((((((((......).))))))))))..))...(((.((((..(((((((((((.(((........)))..))))))))))).))))))))))... ( -31.30) >DroEre_CAF1 12636 107 + 1 GGAAUGACUCAAUUUAUUCGUAUGGGAAUUUAAUUGACACAAAACUGGCGAAACAGAAUGCGAAAAGGAUUAACAUUUUCUAUUUCGCAUUUGUUUUAGAAUCCACC (((.((..((((((.((((......))))..))))))..))........((((((((.(((((((((((........)))).)))))))))))))))....)))... ( -25.70) >DroYak_CAF1 12559 107 + 1 GCAAUGACUCAGUUUAUUUGCAUGGGAAUUUAAUUGACACAAAACUGGCAAAACCGAAUGCGAAAAGGAUAAACAUUUUCCAUUUCGCAUUUGUUUUGGAAUGCACC (((.....(((((((...((((............)).))..)))))))((((((.((((((((((.(((.........))).))))))))))))))))...)))... ( -28.30) >consensus GGAAUGACUCAAUUUAAUCGCAUGGGAAUUUAAUUGACACAAAACUGACAAAACCAAAUGCGAAAAGGAUUAACAUUUCCCAUUUCGCAUUUGUUUUGCAGUCCAGC (((.((..((((((...((......))....))))))..)).......((((((.((((((((((.((...........)).))))))))))))))))...)))... (-19.02 = -18.98 + -0.04)

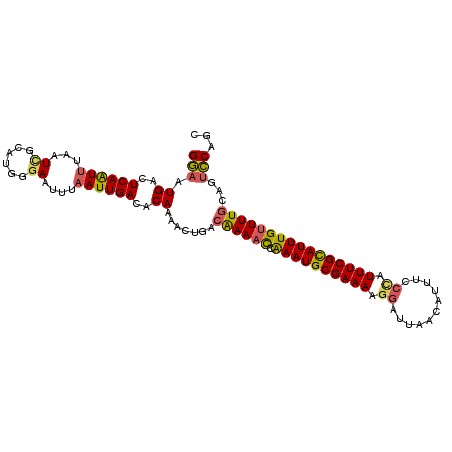

| Location | 12,771,927 – 12,772,035 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.14 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12771927 108 + 23771897 CACAAAACUGACAAAACCAAAUGCGAAAAGGAUUAACAUUUCAAAUUUCGCAUUUGUUUUGCAGGCCAGCACAAAGGAAUGUCACUCAAGAUGAUAUAAACAUUUUAA .......(((.((((((.((((((((((..((........))...))))))))))))))))))).((........)).(((((((....).))))))........... ( -25.40) >DroSec_CAF1 13895 108 + 1 CACAAAACUGACAAAACCAAAUGCGAAAAGGAUUAACAUUUCCCAUUUCGUAUUUGUUUUGCAGUCCAGCACAAAGGAAUGUCGCUCAAGAUGAUAUAAACAUUUUAA ......((((.((((((.((((((((((.(((........)))..))))))))))))))))))))..((((((......))).))).((((((.......)))))).. ( -27.10) >DroSim_CAF1 13265 108 + 1 CACAAAACUGACAAAAUCAAAUGCGAAAAGGAUUAACAUUUCCCAUUUCGCAUUUGCUUUGCAGUCCAGCACAAAGGAAUGUCGCUCAAGAUGAUAUAAACAUUUUAA ......((((.((((..(((((((((((.(((........)))..))))))))))).))))))))..((((((......))).))).((((((.......)))))).. ( -26.80) >DroEre_CAF1 12673 108 + 1 CACAAAACUGGCGAAACAGAAUGCGAAAAGGAUUAACAUUUUCUAUUUCGCAUUUGUUUUAGAAUCCACCACAAAGAAAUGUCGCUCAAUAUGAUAUAAACAUUUUAA ........(((.((((((((.(((((((((((........)))).))))))))))))))).....))).......(((((((...((.....)).....))))))).. ( -19.90) >DroYak_CAF1 12596 108 + 1 CACAAAACUGGCAAAACCGAAUGCGAAAAGGAUAAACAUUUUCCAUUUCGCAUUUGUUUUGGAAUGCACCAAAAAGUAAUGGCGCUUAAGAUUAUAUAAACAUUUUAA ..((((((.((.....))((((((((((.(((.........))).))))))))))))))))....((.(((........))).))....................... ( -26.20) >consensus CACAAAACUGACAAAACCAAAUGCGAAAAGGAUUAACAUUUCCCAUUUCGCAUUUGUUUUGCAGUCCAGCACAAAGGAAUGUCGCUCAAGAUGAUAUAAACAUUUUAA ........(((((....(((((((((((.((...........)).)))))))))))...(((......)))........)))))...((((((.......)))))).. (-17.18 = -17.14 + -0.04)

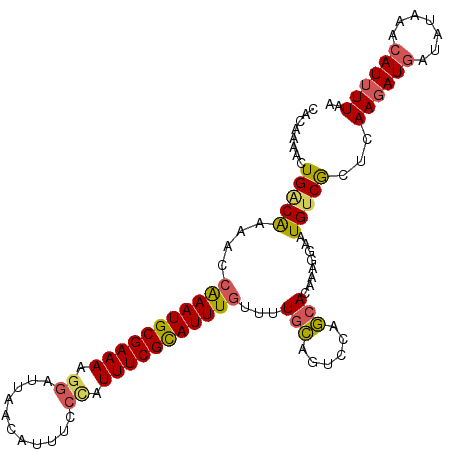
| Location | 12,771,927 – 12,772,035 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -17.86 |
| Energy contribution | -18.46 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12771927 108 - 23771897 UUAAAAUGUUUAUAUCAUCUUGAGUGACAUUCCUUUGUGCUGGCCUGCAAAACAAAUGCGAAAUUUGAAAUGUUAAUCCUUUUCGCAUUUGGUUUUGUCAGUUUUGUG ....((((((....((.....))..)))))).......((..(.(((((((((((((((((((...((........))..)))))))))).)))))).))).)..)). ( -24.00) >DroSec_CAF1 13895 108 - 1 UUAAAAUGUUUAUAUCAUCUUGAGCGACAUUCCUUUGUGCUGGACUGCAAAACAAAUACGAAAUGGGAAAUGUUAAUCCUUUUCGCAUUUGGUUUUGUCAGUUUUGUG ......((((((........))))))(((......)))((..(((((((((((((((.(((((.((((........))))))))).)))).)))))).)))))..)). ( -26.50) >DroSim_CAF1 13265 108 - 1 UUAAAAUGUUUAUAUCAUCUUGAGCGACAUUCCUUUGUGCUGGACUGCAAAGCAAAUGCGAAAUGGGAAAUGUUAAUCCUUUUCGCAUUUGAUUUUGUCAGUUUUGUG ......((((((........))))))(((......)))((..(((((((((((((((((((((.((((........))))))))))))))).))))).)))))..)). ( -33.10) >DroEre_CAF1 12673 108 - 1 UUAAAAUGUUUAUAUCAUAUUGAGCGACAUUUCUUUGUGGUGGAUUCUAAAACAAAUGCGAAAUAGAAAAUGUUAAUCCUUUUCGCAUUCUGUUUCGCCAGUUUUGUG ............(((((((..(((......)))..))))))).........(((((.((((((((((..((((.((.....)).))))))))))))))....))))). ( -23.50) >DroYak_CAF1 12596 108 - 1 UUAAAAUGUUUAUAUAAUCUUAAGCGCCAUUACUUUUUGGUGCAUUCCAAAACAAAUGCGAAAUGGAAAAUGUUUAUCCUUUUCGCAUUCGGUUUUGCCAGUUUUGUG .......................((((((........))))))....((((((.(((((((((.(((.........))).))))))))).((.....)).)))))).. ( -29.70) >consensus UUAAAAUGUUUAUAUCAUCUUGAGCGACAUUCCUUUGUGCUGGACUGCAAAACAAAUGCGAAAUGGGAAAUGUUAAUCCUUUUCGCAUUUGGUUUUGUCAGUUUUGUG .(((((((((((........)))))(((.........(((......)))...(((((((((((.(((.........))).))))))))))).....))).)))))).. (-17.86 = -18.46 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:17 2006