
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,769,381 – 12,769,490 |
| Length | 109 |
| Max. P | 0.983323 |

| Location | 12,769,381 – 12,769,490 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.97 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -21.21 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12769381 109 + 23771897 AUUUGUUCAAUUUA---AUCCA--CUAGGCAUCAUGGUUCUGGCCGUGAUUGGCAUCGUCAUCGGACUCAUUUGCCGCCAGGAUAUUGGCGGCAUCAAAACCAAAUGCUGCCGC ..............---.....--...(((((((((((....)))))))).(((((.(((....))).....(((((((((....)))))))))..........)))))))).. ( -37.30) >DroVir_CAF1 30917 114 + 1 AAAUGUUUCAUAUACGCAUCCAUUGCAGGCAUCAUGGCCGUUGCCAUUGUGGGCAUUAUUGUGGGUUUGAUUUGCCGCAAGGAUGUGGGCGGCAUUAAAACGAAAUGUUGUCGC ..............(((((((.((((.(((((((.(.(((.((((......))))......))).).)))..)))))))))))))))(((((((((.......))))))))).. ( -39.60) >DroEre_CAF1 10119 109 + 1 AUUUGUUCGAUCUU---ACCCA--CCAGGCAUCAUGGUUUUGGCCGUGAUUGGUAUCGUCAUCGGACUCAUUUGCCGCCAGGAUAUUGGCGGCAUCAAAACCAAAUGCUGCCGC ..............---.....--...((((.((((((((((((((((((.......)))).))).......(((((((((....))))))))))))))))))..)).)))).. ( -39.20) >DroYak_CAF1 9986 110 + 1 AU-UGUUCGAUUUU---ACCCACACUAGGCAUCAUGGUUUUGGCCGUGAUUGGCAUCGUCAUCGGACUCAUUUGCCGCCAGGAUAUUGGCGGCAUCAAAACCAAAUGCUGCCGC ..-...........---..........((((.((((((((((((((....))))...(((....))).....(((((((((....))))))))).))))))))..)).)))).. ( -40.10) >DroAna_CAF1 8613 103 + 1 --UUGUUUAAUAUU---UU------CAGGCAUCAUGGUCAUCGCGGUGAUCGGGAUUAUCAUUGGGCUGAUCUGCCGGAAGGACAUUGGAGGGAUCAAGACCAAAUGCUGCCGC --............---..------..((((.(((((((..(.(((((((.......))))))).).((((((.((....)).(......))))))).)))))..)).)))).. ( -29.00) >DroPer_CAF1 36873 97 + 1 --------UACAUU---U------GCAGGCAUCAUGGUAUUGGCGGUCAUUGGCAUUGUCAUUGGCCUCAUUUGCCGCAAGGAUGUGGGCGGCAUCAAGACCAAAUGCUGCCGC --------((((((---(------((.((((..(((((....))(((((.((((...)))).))))).))).)))))))..))))))((((((((.........)))))))).. ( -35.50) >consensus AUUUGUUCAAUAUU___AUCCA__CCAGGCAUCAUGGUUUUGGCCGUGAUUGGCAUCGUCAUCGGACUCAUUUGCCGCAAGGAUAUUGGCGGCAUCAAAACCAAAUGCUGCCGC ...........................(((((((((((....)))))))).(((((.(((....))).....((((((..........))))))..........)))))))).. (-21.21 = -21.80 + 0.59)

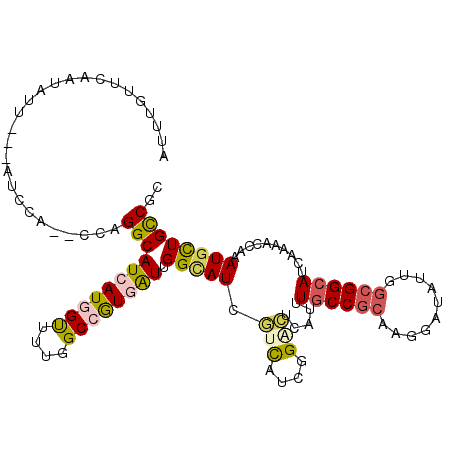
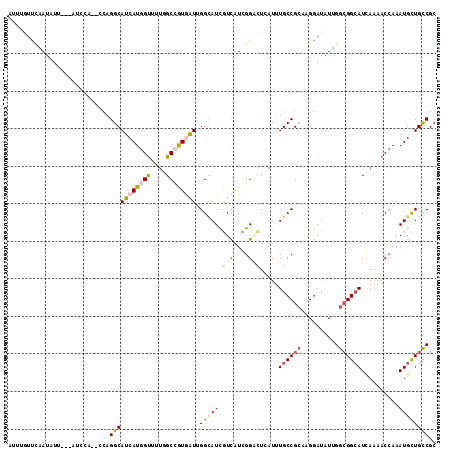
| Location | 12,769,381 – 12,769,490 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.97 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -19.86 |
| Energy contribution | -21.70 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12769381 109 - 23771897 GCGGCAGCAUUUGGUUUUGAUGCCGCCAAUAUCCUGGCGGCAAAUGAGUCCGAUGACGAUGCCAAUCACGGCCAGAACCAUGAUGCCUAG--UGGAU---UAAAUUGAACAAAU ..((((.((..((((((((.((((((((......)))))))).....(((....)))...(((......))))))))))))).))))...--.....---.............. ( -37.40) >DroVir_CAF1 30917 114 - 1 GCGACAACAUUUCGUUUUAAUGCCGCCCACAUCCUUGCGGCAAAUCAAACCCACAAUAAUGCCCACAAUGGCAACGGCCAUGAUGCCUGCAAUGGAUGCGUAUAUGAAACAUUU .............((((((((((.((....((((((((((((.................((....))(((((....)))))..)))).)))).)))))))))).)))))).... ( -27.20) >DroEre_CAF1 10119 109 - 1 GCGGCAGCAUUUGGUUUUGAUGCCGCCAAUAUCCUGGCGGCAAAUGAGUCCGAUGACGAUACCAAUCACGGCCAAAACCAUGAUGCCUGG--UGGGU---AAGAUCGAACAAAU .((.(((((((((((((((.((((((((......)))))))).....(.(((.(((.........))))))))))))))).)))).))).--))...---.............. ( -36.80) >DroYak_CAF1 9986 110 - 1 GCGGCAGCAUUUGGUUUUGAUGCCGCCAAUAUCCUGGCGGCAAAUGAGUCCGAUGACGAUGCCAAUCACGGCCAAAACCAUGAUGCCUAGUGUGGGU---AAAAUCGAACA-AU ..((((.((..((((((((.((((((((......)))))))).....(((....)))...(((......))))))))))))).))))..((.(((..---....))).)).-.. ( -38.10) >DroAna_CAF1 8613 103 - 1 GCGGCAGCAUUUGGUCUUGAUCCCUCCAAUGUCCUUCCGGCAGAUCAGCCCAAUGAUAAUCCCGAUCACCGCGAUGACCAUGAUGCCUG------AA---AAUAUUAAACAA-- ..((((.((..(((((.............((((.....(((......)))....))))(((.((.....)).)))))))))).))))..------..---............-- ( -21.20) >DroPer_CAF1 36873 97 - 1 GCGGCAGCAUUUGGUCUUGAUGCCGCCCACAUCCUUGCGGCAAAUGAGGCCAAUGACAAUGCCAAUGACCGCCAAUACCAUGAUGCCUGC------A---AAUGUA-------- ((((((((((((((((((..((((((..........))))))...))))))).....)))))...)).))))......(((..((....)------)---.)))..-------- ( -28.90) >consensus GCGGCAGCAUUUGGUUUUGAUGCCGCCAAUAUCCUGGCGGCAAAUGAGUCCGAUGACAAUGCCAAUCACGGCCAAAACCAUGAUGCCUGG__UGGAU___AAUAUCGAACAAAU ..((((.((..((((((((.((((((..........))))))..................(((......))))))))))))).))))........................... (-19.86 = -21.70 + 1.84)

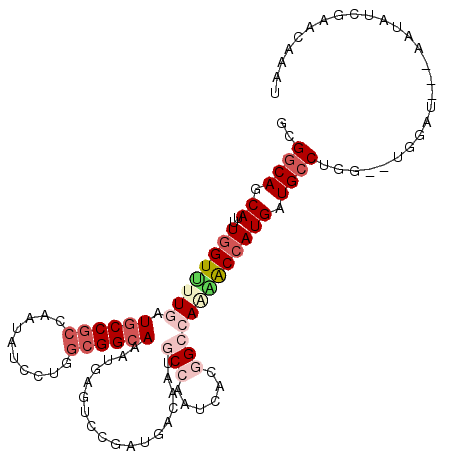
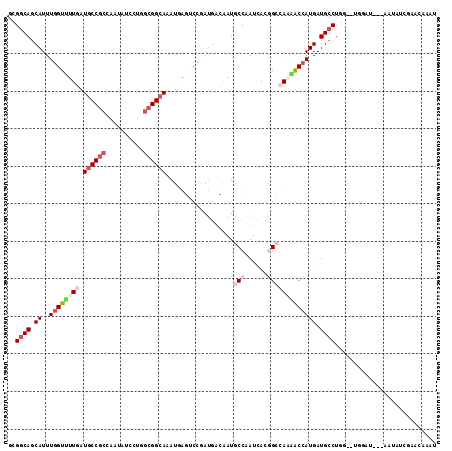
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:13 2006