| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
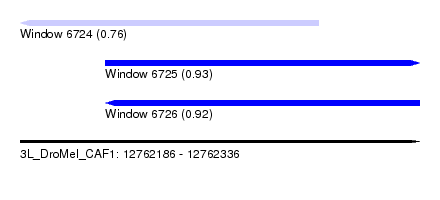
| Location | 12,762,186 – 12,762,336 |
| Length | 150 |
| Max. P | 0.925142 |

| Location | 12,762,186 – 12,762,298 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.59 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
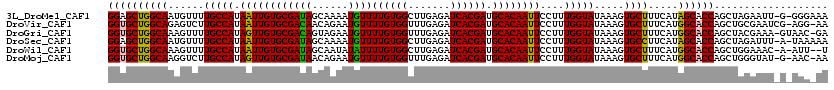
>3L_DroMel_CAF1 12762186 112 - 23771897 ACCAAAGGAAUUGUGCAUCGUGAUCUCAAGCCACAAAACAUUUUGCUAUCGCACAAUUAUGGCAAAACAUUGCCAGCUCCAUCGAAAAUAACCCUGAAAAUUGGUAAGUUUU------- ((((((((((((((((....((....))(((.............)))...)))))))).((((((....)))))).................))).....))))).......------- ( -22.82) >DroVir_CAF1 21980 118 - 1 ACCAAAGGAAUUGUGCAUCGUGAUCUCAAACCACAAAACAUUCUGUUGUCGCACAAUUAUGGCAAGACUCUGCCAGCACCAUCCAAAAUAACGCUGAAAAUUGGUAAGCAUA-UAGGAA .((...((((((((((...(((.........)))...(((......))).)))))))).(((((......)))))...))............(((...........)))...-..)).. ( -25.30) >DroGri_CAF1 15104 117 - 1 ACCAAAGGAAUUGUGCAUCGUGAUCUCAAACCACAAAACAUUCUACUGUCGCACAACUAUGGCAAAACUUUGCCAGCACCAUCGAAAAUAACGCUGAAAAUUGGUAAGCAAA-UCC-AA ......(((.((((((...(((.........)))...(((......))).))))))...((((((....))))))((((((....................))))..))...-)))-.. ( -22.65) >DroWil_CAF1 3389 118 - 1 ACCAAAGGAAUUGUGCAUCGUGAUCUCAAGCCACAAAAUAUAUUGCUAUCGCACAAUUAUGGCAAAACUUUGCCAGCACCAUCGAAAAUAACAUUGAAAAUUGGUAAGUCAAGUCG-CC (((((.((((((((((....((....))(((.............)))...)))))))).((((((....))))))...)).((((........))))...)))))...........-.. ( -24.22) >DroMoj_CAF1 13872 108 - 1 ACCAAAGGAAUUGUGCAUCGUGAUCUCAAACCACAAAACAUUCUGUUAUCGCACAACUAUGGCAAGACCUUGCCAGCACCAUCCAAAAUUACAUUGAAAAUUGGUAAG----------- (((((.(((...((((...(((((..((...............))..))))).......((((((....))))))))))..)))..........(....))))))...----------- ( -24.16) >DroAna_CAF1 2678 117 - 1 ACCAAAGGAAUUGUGCAUCGUGAUCUCAAGCCACAAAAUAUUUUGCUAUCGCACAAUUAUGGCAAAACCUUGCCAGCUCCAUCGAAAAUAACUCUGAAAAUUGGUAAGUAAA-UCA-UC (((((.((((((((((....((....))(((.............)))...)))))))..((((((....))))))..))).((((.......)).))...))))).......-...-.. ( -22.22) >consensus ACCAAAGGAAUUGUGCAUCGUGAUCUCAAACCACAAAACAUUCUGCUAUCGCACAAUUAUGGCAAAACUUUGCCAGCACCAUCGAAAAUAACACUGAAAAUUGGUAAGUAAA_UC____ (((((...((((((((...(((.........)))...(((......))).)))))))).((((((....)))))).........................))))).............. (-19.91 = -20.52 + 0.61)



| Location | 12,762,218 – 12,762,336 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.26 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -31.44 |
| Energy contribution | -30.83 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12762218 118 + 23771897 GGAGCUGGCAAUGUUUUGCCAUAAUUGUGCGAUAGCAAAAUGUUUUGUGGCUUGAGAUCACGAUGCACAAUUCCUUUGGUAUAAAGUGCUUUCAUAGCACCAGCUAGAAUU-G-GGGAAA ..((((((...((((..(((((((...(((....))).......))))))).(((((.(((.((((.(((.....)))))))...))).))))).))))))))))......-.-...... ( -32.90) >DroVir_CAF1 22018 118 + 1 GGUGCUGGCAGAGUCUUGCCAUAAUUGUGCGACAACAGAAUGUUUUGUGGUUUGAGAUCACGAUGCACAAUUCCUUUGGUAUAAAGUGCUUUCAUGGCACCAGCUGCGAAUCG-AGG-AA (((((((..((((((((((((.((((((((((((......))))((((((((...)))))))).))))))))....)))))...)).)))))..)))))))............-...-.. ( -34.50) >DroGri_CAF1 15141 118 + 1 GGUGCUGGCAAAGUUUUGCCAUAGUUGUGCGACAGUAGAAUGUUUUGUGGUUUGAGAUCACGAUGCACAAUUCCUUUGGUAUAAAGUGCUUUCAUGGCACCAGCUACGAAA-GUAAC-GA ((((((((((...((.(((((.((((((((((((......))))((((((((...)))))))).))))))))....))))).))..)))).....)))))).(.(((....-))).)-.. ( -40.40) >DroSec_CAF1 4131 118 + 1 GGAGCUGGCAAUGUUUUGCCAUAAUUGUGCGAUAGCAAAAUGUUUUGUGGCUUGAGAUCACGAUGCACAAUUCCUUUGGUAUAAAGUGCCUUCAUAGCACCAGCUAGAUUU-A-UAAAAA ..((((((...((((..(((((((...(((....))).......))))))).((((..(((.((((.(((.....)))))))...)))..)))).))))))))))......-.-...... ( -30.90) >DroWil_CAF1 3427 116 + 1 GGUGCUGGCAAAGUUUUGCCAUAAUUGUGCGAUAGCAAUAUAUUUUGUGGCUUGAGAUCACGAUGCACAAUUCCUUUGGUAUAAAGUGCUUUCAUGGCACCAGCUGGAAAC-A-AUU--U ((((((((((...((.(((((.((((((((((((....)))...(((((((....).)))))))))))))))....))))).))..)))).....))))))...((....)-)-...--. ( -36.20) >DroMoj_CAF1 13900 117 + 1 GGUGCUGGCAAGGUCUUGCCAUAGUUGUGCGAUAACAGAAUGUUUUGUGGUUUGAGAUCACGAUGCACAAUUCCUUUGGUAUAAAGUGCUUUCAUGGCACCAGCUGGGUAU-G-AAC-AA ((((((((((...(..(((((.(((((((((..(((.....)))((((((((...)))))))))))))))))....)))))..)..)))).....))))))..........-.-...-.. ( -34.10) >consensus GGUGCUGGCAAAGUUUUGCCAUAAUUGUGCGAUAGCAAAAUGUUUUGUGGCUUGAGAUCACGAUGCACAAUUCCUUUGGUAUAAAGUGCUUUCAUGGCACCAGCUAGAAAU_G_AAC_AA ((((((((((......(((((.((((((((((((......))))((((((.......)))))).))))))))....))))).....)))).....))))))................... (-31.44 = -30.83 + -0.61)


| Location | 12,762,218 – 12,762,336 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.26 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -24.75 |
| Energy contribution | -24.81 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12762218 118 - 23771897 UUUCCC-C-AAUUCUAGCUGGUGCUAUGAAAGCACUUUAUACCAAAGGAAUUGUGCAUCGUGAUCUCAAGCCACAAAACAUUUUGCUAUCGCACAAUUAUGGCAAAACAUUGCCAGCUCC ......-.-......((((((((((((((..((((...((.((...)).)).))))...(((((..((((...........))))..)))))....)))))))........))))))).. ( -26.20) >DroVir_CAF1 22018 118 - 1 UU-CCU-CGAUUCGCAGCUGGUGCCAUGAAAGCACUUUAUACCAAAGGAAUUGUGCAUCGUGAUCUCAAACCACAAAACAUUCUGUUGUCGCACAAUUAUGGCAAGACUCUGCCAGCACC ..-...-.........(((((((((((((.....((((.....))))...((((((...(((.........)))...(((......))).)))))))))))))........))))))... ( -28.50) >DroGri_CAF1 15141 118 - 1 UC-GUUAC-UUUCGUAGCUGGUGCCAUGAAAGCACUUUAUACCAAAGGAAUUGUGCAUCGUGAUCUCAAACCACAAAACAUUCUACUGUCGCACAACUAUGGCAAAACUUUGCCAGCACC ..-....(-(((.(((...(((((.......)))))...))).))))...((((((...(((.........)))...(((......))).))))))...((((((....))))))..... ( -27.40) >DroSec_CAF1 4131 118 - 1 UUUUUA-U-AAAUCUAGCUGGUGCUAUGAAGGCACUUUAUACCAAAGGAAUUGUGCAUCGUGAUCUCAAGCCACAAAACAUUUUGCUAUCGCACAAUUAUGGCAAAACAUUGCCAGCUCC ......-.-......((((((((((((((..((((...((.((...)).)).))))...(((((..((((...........))))..)))))....)))))))........))))))).. ( -26.00) >DroWil_CAF1 3427 116 - 1 A--AAU-U-GUUUCCAGCUGGUGCCAUGAAAGCACUUUAUACCAAAGGAAUUGUGCAUCGUGAUCUCAAGCCACAAAAUAUAUUGCUAUCGCACAAUUAUGGCAAAACUUUGCCAGCACC .--...-.-.......(((((((((((((..((((...((.((...)).)).))))...(((((..(((.............)))..)))))....)))))))........))))))... ( -28.62) >DroMoj_CAF1 13900 117 - 1 UU-GUU-C-AUACCCAGCUGGUGCCAUGAAAGCACUUUAUACCAAAGGAAUUGUGCAUCGUGAUCUCAAACCACAAAACAUUCUGUUAUCGCACAACUAUGGCAAGACCUUGCCAGCACC ..-...-.-.......((((((((((((......((((.....))))...((((((...(((.........)))..(((.....)))...)))))).))))))........))))))... ( -26.80) >consensus UU_CUU_C_AUUCCCAGCUGGUGCCAUGAAAGCACUUUAUACCAAAGGAAUUGUGCAUCGUGAUCUCAAACCACAAAACAUUCUGCUAUCGCACAAUUAUGGCAAAACUUUGCCAGCACC ................(((((((((((((..((((...((.((...)).)).))))...(((((..((...............))..)))))....)))))))........))))))... (-24.75 = -24.81 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:10 2006