
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,758,808 – 12,758,957 |
| Length | 149 |
| Max. P | 0.990672 |

| Location | 12,758,808 – 12,758,918 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.48 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -15.34 |
| Energy contribution | -14.86 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12758808 110 - 23771897 AUAUAGUCGUAAGAUAACCAUAAUAU---UAUAUAUAUGUUUACAUAUAUGU--ACAACACGAAAGCUAACCAAGCAAACCAC----AUUACA-UAACGUAAAGGGCAAUAUGCAAGCCA .....(((....)))...........---((((((((((....)))))))))--)..........(((.....)))......(----.((((.-....)))).)(((.........))). ( -17.90) >DroSim_CAF1 18057 109 - 1 AUAUAGUCGUAAGAUAACCAUAAUAU---UAUAUAUAUGUUUACAUAUAUAU--AU-ACACGAAAGCUAACCAAGCAAACCAC----AUUACA-UAACGUAAAGGGCAAUAUGCAAGCCA .....(((....)))...........---((((((((((....)))))))))--).-........(((.....)))......(----.((((.-....)))).)(((.........))). ( -18.00) >DroEre_CAF1 15978 109 - 1 AUAUAGUCGUAAGAUAACCAUAA------UAUAUAUAUGUUAACAUAUAUAUGUACAACACGAAAGCUAACCAAGCAAACAAC----AUUACA-UAACGUAAAGGGCAAUAUGCAAGCCA .....(((....))).......(------((((((((((....)))))))))))...........(((.....)))......(----.((((.-....)))).)(((.........))). ( -19.30) >DroYak_CAF1 16366 112 - 1 AUAUAGUCGUAAGAUAACCAUAAAAU---UAUAUAUAUGUUAACAUAUAUAUGUACAACACGAAAGCUAACCAAGCAAACCAC----AUUACA-UAACGUAAAGGGCAAUAUGCAAGCCA .....(((....)))...........---((((((((((....))))))))))............(((.....)))......(----.((((.-....)))).)(((.........))). ( -17.70) >DroAna_CAF1 20110 119 - 1 AUGUAGAUCUAAGAUAACCAUAAUAUAUAAAUAUAUAUGUUUACAUAUAUAUGUGCAACACGAAAGCUAACCAAGCAA-CCAUAUGGAUUAGAGUAACAUAAAGGGCAAUAUGCAAGCCA ((((...(((((.....(((((........(((((((((....))))))))).............(((.....)))..-...))))).)))))...))))....(((.........))). ( -23.20) >consensus AUAUAGUCGUAAGAUAACCAUAAUAU___UAUAUAUAUGUUUACAUAUAUAUGUACAACACGAAAGCUAACCAAGCAAACCAC____AUUACA_UAACGUAAAGGGCAAUAUGCAAGCCA ........((((..................(((((((((....))))))))).............(((.....)))............))))............(((.........))). (-15.34 = -14.86 + -0.48)

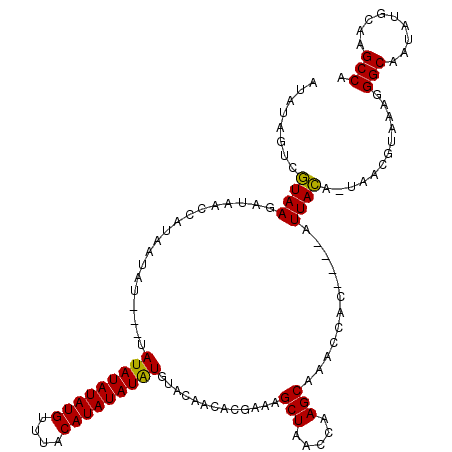

| Location | 12,758,843 – 12,758,957 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.57 |
| Mean single sequence MFE | -22.01 |
| Consensus MFE | -18.10 |
| Energy contribution | -17.82 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12758843 114 - 23771897 GAGUGCAGAUGCAACAUGAAUAAGGGCAGACAA-GAGUCCAUAUAGUCGUAAGAUAACCAUAAUAU---UAUAUAUAUGUUUACAUAUAUGU--ACAACACGAAAGCUAACCAAGCAAAC ..(((((.(((.((((((.(((.((((......-..)))).....(((....)))...........---..))).))))))..)))...)))--)).........(((.....))).... ( -22.00) >DroSim_CAF1 18092 113 - 1 GAGUGCAGAUGCAACAUGAAUAAGGGCAGACAA-GAGUCCAUAUAGUCGUAAGAUAACCAUAAUAU---UAUAUAUAUGUUUACAUAUAUAU--AU-ACACGAAAGCUAACCAAGCAAAC ..(((..........(((.....((((......-..)))).....(((....)))...))).....---((((((((((....)))))))))--).-.)))....(((.....))).... ( -21.60) >DroEre_CAF1 16013 113 - 1 GAGUGCAGAUGCAACAUGAAUAAGGGCGGAUAA-GAGUCCAUAUAGUCGUAAGAUAACCAUAA------UAUAUAUAUGUUAACAUAUAUAUGUACAACACGAAAGCUAACCAAGCAAAC ..(((..................((..((((..-..)))).....(((....)))..))...(------((((((((((....)))))))))))....)))....(((.....))).... ( -23.70) >DroYak_CAF1 16401 116 - 1 GAGUGCAGAUGCAACAUGAAUAAGGGCAGAUAA-GAGUCCAUAUAGUCGUAAGAUAACCAUAAAAU---UAUAUAUAUGUUAACAUAUAUAUGUACAACACGAAAGCUAACCAAGCAAAC ..((((.........(((.....((((......-..)))).....(((....)))...))).....---.(((((((((....))))))))))))).........(((.....))).... ( -22.60) >DroAna_CAF1 20150 119 - 1 GAGUGCAGAUGCAACAUGAAUAAGGGCAGAUAAGGAGUCCAUGUAGAUCUAAGAUAACCAUAAUAUAUAAAUAUAUAUGUUUACAUAUAUAUGUGCAACACGAAAGCUAACCAAGCAA-C ...(((.(.((((.((((.....((((.........))))((((((((..............((((((....))))))))))))))...)))))))).).(....)........))).-. ( -20.13) >consensus GAGUGCAGAUGCAACAUGAAUAAGGGCAGAUAA_GAGUCCAUAUAGUCGUAAGAUAACCAUAAUAU___UAUAUAUAUGUUUACAUAUAUAUGUACAACACGAAAGCUAACCAAGCAAAC ..(((..........(((.(((.((((.........)))).))).(((....)))...))).........(((((((((....)))))))))......)))....(((.....))).... (-18.10 = -17.82 + -0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:06 2006