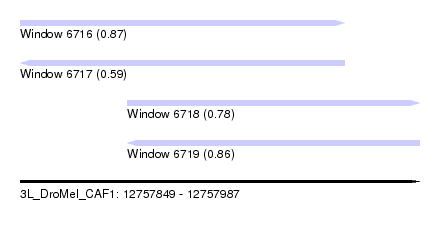
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,757,849 – 12,757,987 |
| Length | 138 |
| Max. P | 0.871983 |

| Location | 12,757,849 – 12,757,961 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -33.58 |
| Energy contribution | -34.20 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
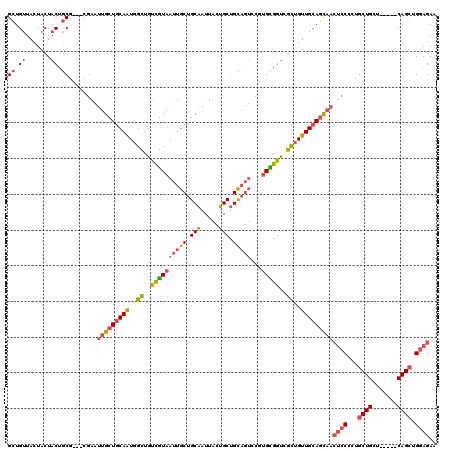
>3L_DroMel_CAF1 12757849 112 + 23771897 GCUGUUACUACUACUGCG---CGAAUUGCUGCAAUGGCUGUCGUAAUUGCUGCAAUUACUGCUGCAGUCCGUGCGGUUGCUGUUGCAGCAACUCCCCUGCUGCU-----CAGCUGGAGAA ((.((.......)).)).---....(((((((((((((..(((((...((((((........))))))...)))))..)))))))))))))((((...(((...-----.))).)))).. ( -45.80) >DroSec_CAF1 14806 112 + 1 GCUGUUACUACUACUGCG---CGAAUUGCUGCAAUGGCUGUCGUAAUUGCUGCAAUUACUGCUGCAGUCCGUGCGGUCGCUGUUGCAGCAACUCCCCUGCUGCU-----CAGCUGGAGAA ((.((.......)).)).---....(((((((((((((..(((((...((((((........))))))...)))))..)))))))))))))((((...(((...-----.))).)))).. ( -45.70) >DroSim_CAF1 17089 112 + 1 GCUGUUACUACUACUGCG---CGAAUUGCUGCAAUGGCUGUCGUAAUUGCUGCAAUUACUGCUGCAGUCCGUGCGGUCGCUGUUGCAGCAACUCCCCUGCUGCU-----CAGCUGGAGAA ((.((.......)).)).---....(((((((((((((..(((((...((((((........))))))...)))))..)))))))))))))((((...(((...-----.))).)))).. ( -45.70) >DroEre_CAF1 15035 112 + 1 GCUGCUACUACUACUGCG---CGAAUUGCUGCAAUGGCUGUCGUAAUUGCUGCAAUUACUGCUGUAGUCCGGGCGGUCGCUGUUGCAGCAACUCCGCUGCUGCU-----CAGCUGGAGAA ((((((...(((((.(((---..((((((.(((((.((....)).))))).))))))..))).)))))...)))))).((((..((((((.......)))))).-----))))....... ( -46.30) >DroYak_CAF1 15250 112 + 1 GCUGUUACUACUACUGCG---CGAAUUGCUGCAAUGGCUGUCGUAAUUGCUGCAAUUACUGCUGUAGUCCGUGUGGUCGCUGUUGCAGUAACUCCCAUGCUGCU-----CAGCUGGAGAA (((((.((..(.((..((---((.(((((.(((.(((.(((.((....)).))).))).))).))))).))))..)).)..)).)))))..((((...(((...-----.))).)))).. ( -38.40) >DroAna_CAF1 19097 110 + 1 GAUGUUAUUACUACUACCCUUCAAGUUGUUACAGUUGUUGCUAUAGUUGCUGCUAUCCCGGCUGU------UGUGGCAGUUGUUGCAUC----CCCAAGCUGCCACCGCCAGAUGCCGAU (((((....((.(((........))).)).))(((.((.((....)).)).)))))).((((.((------.(((((((((........----....))))))))).)).....)))).. ( -26.00) >consensus GCUGUUACUACUACUGCG___CGAAUUGCUGCAAUGGCUGUCGUAAUUGCUGCAAUUACUGCUGCAGUCCGUGCGGUCGCUGUUGCAGCAACUCCCCUGCUGCU_____CAGCUGGAGAA ((.((.......)).))........(((((((((..((..((((((((((.(((.....))).)))))...)))))..))..)))))))))((((...((((.......)))).)))).. (-33.58 = -34.20 + 0.62)

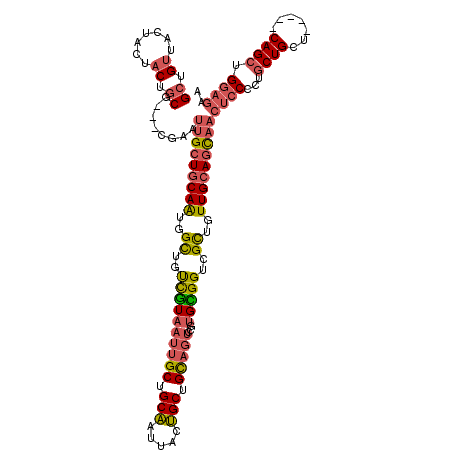
| Location | 12,757,849 – 12,757,961 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -25.64 |
| Energy contribution | -27.37 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12757849 112 - 23771897 UUCUCCAGCUG-----AGCAGCAGGGGAGUUGCUGCAACAGCAACCGCACGGACUGCAGCAGUAAUUGCAGCAAUUACGACAGCCAUUGCAGCAAUUCG---CGCAGUAGUAGUAACAGC .(((((.(((.-----...))).)))))((((((((........((....))(((((.((.(.((((((.(((((..(....)..))))).))))))))---)))))).))))))))... ( -41.60) >DroSec_CAF1 14806 112 - 1 UUCUCCAGCUG-----AGCAGCAGGGGAGUUGCUGCAACAGCGACCGCACGGACUGCAGCAGUAAUUGCAGCAAUUACGACAGCCAUUGCAGCAAUUCG---CGCAGUAGUAGUAACAGC .(((((.(((.-----...))).)))))((((((((....((....))....(((((.((.(.((((((.(((((..(....)..))))).))))))))---)))))).))))))))... ( -42.00) >DroSim_CAF1 17089 112 - 1 UUCUCCAGCUG-----AGCAGCAGGGGAGUUGCUGCAACAGCGACCGCACGGACUGCAGCAGUAAUUGCAGCAAUUACGACAGCCAUUGCAGCAAUUCG---CGCAGUAGUAGUAACAGC .(((((.(((.-----...))).)))))((((((((....((....))....(((((.((.(.((((((.(((((..(....)..))))).))))))))---)))))).))))))))... ( -42.00) >DroEre_CAF1 15035 112 - 1 UUCUCCAGCUG-----AGCAGCAGCGGAGUUGCUGCAACAGCGACCGCCCGGACUACAGCAGUAAUUGCAGCAAUUACGACAGCCAUUGCAGCAAUUCG---CGCAGUAGUAGUAGCAGC .......((((-----.((.((.(((((((((((((((..((..((....))......(..(((((((...)))))))..).))..)))))))))))).---))).)).))..))))... ( -40.20) >DroYak_CAF1 15250 112 - 1 UUCUCCAGCUG-----AGCAGCAUGGGAGUUACUGCAACAGCGACCACACGGACUACAGCAGUAAUUGCAGCAAUUACGACAGCCAUUGCAGCAAUUCG---CGCAGUAGUAGUAACAGC .((.((((((.-----...))).)))))((((((((.((.((..((....))......((.(.((((((.(((((..(....)..))))).))))))))---))).)).))))))))... ( -34.90) >DroAna_CAF1 19097 110 - 1 AUCGGCAUCUGGCGGUGGCAGCUUGGG----GAUGCAACAACUGCCACA------ACAGCCGGGAUAGCAGCAACUAUAGCAACAACUGUAACAACUUGAAGGGUAGUAGUAAUAACAUC ....((.((((((.(((((((.(((..----.......)))))))))).------...))))))...))....(((((.((.......((....)).......)).)))))......... ( -29.24) >consensus UUCUCCAGCUG_____AGCAGCAGGGGAGUUGCUGCAACAGCGACCGCACGGACUACAGCAGUAAUUGCAGCAAUUACGACAGCCAUUGCAGCAAUUCG___CGCAGUAGUAGUAACAGC .(((((.((((.......)))).)))))((((((((.((.(((.((....)).........(.((((((.(((((..(....)..))))).)))))))....))).)).))))))))... (-25.64 = -27.37 + 1.73)

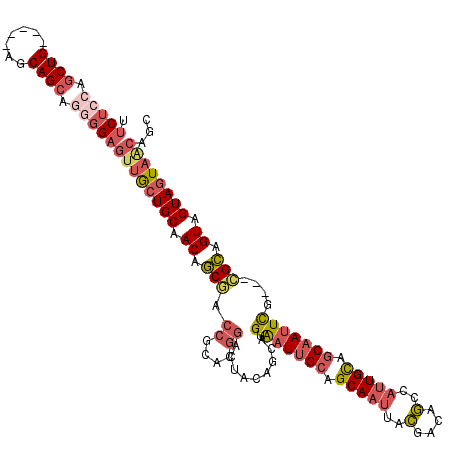

| Location | 12,757,886 – 12,757,987 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12757886 101 + 23771897 UCGUAAUUGCUGCAAUUACUGCUGCAGUCCGUGCGGUUGCUGUUGCAGCAACUCCCCUGCUGCU-----CAGCUGGAGAAG-----AUUCCUCCGA----AGCCGAAGCAAAAGG----- ......((((((((((..(.((((((.....)))))).)..))))))))))....(((..((((-----(.(((((((...-----....))))..----))).).))))..)))----- ( -38.20) >DroSec_CAF1 14843 101 + 1 UCGUAAUUGCUGCAAUUACUGCUGCAGUCCGUGCGGUCGCUGUUGCAGCAACUCCCCUGCUGCU-----CAGCUGGAGAAG-----AUUCCUCCGA----AGCCGAAGCAAAAGG----- ......((((((((((..(.((((((.....)))))).)..))))))))))....(((..((((-----(.(((((((...-----....))))..----))).).))))..)))----- ( -38.20) >DroSim_CAF1 17126 101 + 1 UCGUAAUUGCUGCAAUUACUGCUGCAGUCCGUGCGGUCGCUGUUGCAGCAACUCCCCUGCUGCU-----CAGCUGGAGAAG-----AUUCCUCCGA----AGCCGAAGCAAAAGG----- ......((((((((((..(.((((((.....)))))).)..))))))))))....(((..((((-----(.(((((((...-----....))))..----))).).))))..)))----- ( -38.20) >DroEre_CAF1 15072 101 + 1 UCGUAAUUGCUGCAAUUACUGCUGUAGUCCGGGCGGUCGCUGUUGCAGCAACUCCGCUGCUGCU-----CAGCUGGAGAAG-----AUCCCUCCGA----AGCCGAAGCAAAAGG----- ......(((((.(.(((((....)))))...(((..((((((..((((((.......)))))).-----)))).((((...-----....))))))----.)))).)))))....----- ( -34.90) >DroYak_CAF1 15287 101 + 1 UCGUAAUUGCUGCAAUUACUGCUGUAGUCCGUGUGGUCGCUGUUGCAGUAACUCCCAUGCUGCU-----CAGCUGGAGAAG-----AUUACUCCAA----AGCCGAAGCAAAAGG----- ......(((((((((((((....)))))...)))(((.((((..((((((.......)))))).-----))))(((((...-----....))))).----.)))..)))))....----- ( -29.40) >DroAna_CAF1 19137 110 + 1 CUAUAGUUGCUGCUAUCCCGGCUGU------UGUGGCAGUUGUUGCAUC----CCCAAGCUGCCACCGCCAGAUGCCGAUGUCACCAUACCACCAAAAAAAUCCAAAGAUAAAGAUAAAG ..(((((....)))))..((((.((------.(((((((((........----....))))))))).)).....)))).......................................... ( -23.60) >consensus UCGUAAUUGCUGCAAUUACUGCUGCAGUCCGUGCGGUCGCUGUUGCAGCAACUCCCCUGCUGCU_____CAGCUGGAGAAG_____AUUCCUCCGA____AGCCGAAGCAAAAGG_____ ......((((((((((..(.((((((.....)))))).)..))))))))))((((...((((.......)))).)))).......................((....))........... (-24.38 = -25.02 + 0.64)


| Location | 12,757,886 – 12,757,987 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -23.28 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12757886 101 - 23771897 -----CCUUUUGCUUCGGCU----UCGGAGGAAU-----CUUCUCCAGCUG-----AGCAGCAGGGGAGUUGCUGCAACAGCAACCGCACGGACUGCAGCAGUAAUUGCAGCAAUUACGA -----(((.(((((.(((((----..((((((..-----.)))))))))))-----))))).)))..(((((((((((..((..(.(((.....))).)..))..))))))))))).... ( -40.10) >DroSec_CAF1 14843 101 - 1 -----CCUUUUGCUUCGGCU----UCGGAGGAAU-----CUUCUCCAGCUG-----AGCAGCAGGGGAGUUGCUGCAACAGCGACCGCACGGACUGCAGCAGUAAUUGCAGCAAUUACGA -----(((.(((((.(((((----..((((((..-----.)))))))))))-----))))).)))..(((((((((((..((..(.(((.....))).)..))..))))))))))).... ( -41.60) >DroSim_CAF1 17126 101 - 1 -----CCUUUUGCUUCGGCU----UCGGAGGAAU-----CUUCUCCAGCUG-----AGCAGCAGGGGAGUUGCUGCAACAGCGACCGCACGGACUGCAGCAGUAAUUGCAGCAAUUACGA -----(((.(((((.(((((----..((((((..-----.)))))))))))-----))))).)))..(((((((((((..((..(.(((.....))).)..))..))))))))))).... ( -41.60) >DroEre_CAF1 15072 101 - 1 -----CCUUUUGCUUCGGCU----UCGGAGGGAU-----CUUCUCCAGCUG-----AGCAGCAGCGGAGUUGCUGCAACAGCGACCGCCCGGACUACAGCAGUAAUUGCAGCAAUUACGA -----.((.(((((.(((((----..(((((...-----..))))))))))-----))))).))((.(((((((((((......((....))..(((....))).))))))))))).)). ( -35.70) >DroYak_CAF1 15287 101 - 1 -----CCUUUUGCUUCGGCU----UUGGAGUAAU-----CUUCUCCAGCUG-----AGCAGCAUGGGAGUUACUGCAACAGCGACCACACGGACUACAGCAGUAAUUGCAGCAAUUACGA -----.....((((.(((((----..((((....-----...)))))))))-----))))((...(.(((((((((...((...((....)).))...))))))))).).))........ ( -30.40) >DroAna_CAF1 19137 110 - 1 CUUUAUCUUUAUCUUUGGAUUUUUUUGGUGGUAUGGUGACAUCGGCAUCUGGCGGUGGCAGCUUGGG----GAUGCAACAACUGCCACA------ACAGCCGGGAUAGCAGCAACUAUAG ........(((((..............((.(.(((....)))).))..(((((.(((((((.(((..----.......)))))))))).------...))))))))))............ ( -32.10) >consensus _____CCUUUUGCUUCGGCU____UCGGAGGAAU_____CUUCUCCAGCUG_____AGCAGCAGGGGAGUUGCUGCAACAGCGACCGCACGGACUACAGCAGUAAUUGCAGCAAUUACGA ...........((....)).......((((((........)))))).((((......(((((((.....)))))))........((....))....)))).(((((((...))))))).. (-23.28 = -24.23 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:03 2006