| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
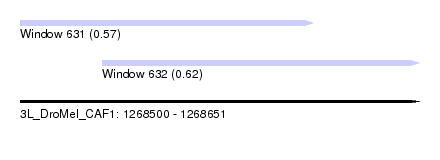
| Location | 1,268,500 – 1,268,651 |
| Length | 151 |
| Max. P | 0.624786 |

| Location | 1,268,500 – 1,268,611 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -37.71 |
| Consensus MFE | -26.43 |
| Energy contribution | -26.85 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
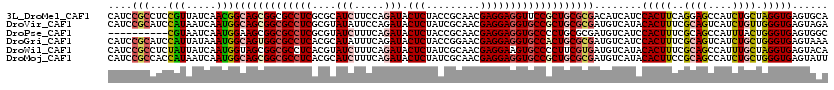
>3L_DroMel_CAF1 1268500 111 + 23771897 CUCCACGAAUCACCGCC------GGG---GGGCAGCUGCUCAUCCGCCUCCGUUAUCAACGGCAGCGGCGCCUCGCGCAUCUUCCAGAUACUCUACCGCAACGAGGAGGUUCCGCUGCGC ..............(.(------(((---((((....)))).)))).)..(((.....)))(((((((.(((((....(((.....))).(((.........)))))))).))))))).. ( -36.80) >DroVir_CAF1 25929 103 + 1 --------AG---UGCC------GGGCAUCGGCAGCUGCACAUCCGCAUCCAUAAUCAAUGGCAGCGGCGCCUCGCGUAUAUUCCAGAUACUCUAUCGCAACGAGGAGGUGCCGCUGCGC --------..---((((------(.....)))))(((((......))).............(((((((((((((.(((.......((.....))......)))..))))))))))))))) ( -40.82) >DroGri_CAF1 36314 103 + 1 --------AA---CGCU------GGGCAGCGGCAGUUGUACAUCCGCAUCCAUUAUAAAUGGCAGUGGCGCCUCACGCAUAUUUCAGAUACUCUACCGGAACGAGGAGGUGCCACUGCGC --------..---.(((------(((..((((..(.....)..)))).)))).........(((((((((((((.....((((...))))(((.........)))))))))))))))))) ( -36.60) >DroWil_CAF1 34595 108 + 1 ---------UCAUCGUCAGGACUGCC---AGGUAGCUGUACAUCCGCCUCUAUUAUCAAUGGUAGCGGCGCCUCACGUAUCUUUCAGAUACUCUAUCGCAACGAGGAAGUGCCCCUUCGU ---------....((..(((.((((.---..))))..((((.((((((.((((((....)))))).)))((.....(((((.....)))))......)).....))).)))).))).)). ( -27.10) >DroMoj_CAF1 27107 103 + 1 --------CG---UGCC------GGGCAUUGGCAGCUGCACAUCCGCCACCAUAAUCAAUGGCAGCGGCGCCUCACGCAUCUUUCAGAUACUCUAUCGCAACGAGGAGGUGCCGCUGCGC --------.(---((.(------(((...((((....)).)))))).)))...........(((((((((((((....(((.....))).(((.........)))))))))))))))).. ( -41.20) >DroAna_CAF1 24141 111 + 1 CUGGGCGGGUCCUCCCU------GCC---GGGCAGCUGCUCAUCCGCCUCGGUUAUCAACGGGAGCGGCGCCUCGCGCAUCUUUCAGAUACUCUACCGCAACGAGGAGGUUCCGCUGCGC ...(((((..((((((.------(((---((((.(((((((..(((.............))))))))))))).)).))(((.....))).............).)))))..))))).... ( -43.72) >consensus ________AU___CGCC______GGG___CGGCAGCUGCACAUCCGCCUCCAUUAUCAAUGGCAGCGGCGCCUCACGCAUCUUUCAGAUACUCUACCGCAACGAGGAGGUGCCGCUGCGC ...............................(((((.........((..((((.....))))..))((((((((....(((.....))).(((.........)))))))))))))))).. (-26.43 = -26.85 + 0.42)
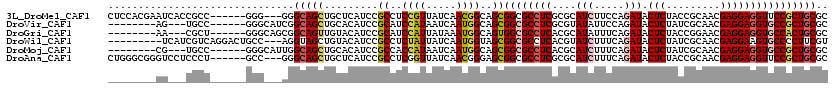
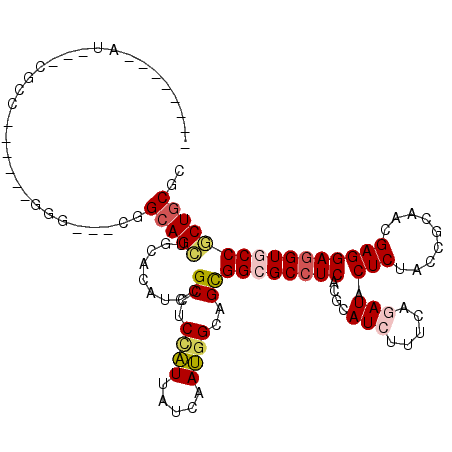
| Location | 1,268,531 – 1,268,651 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -31.12 |
| Energy contribution | -31.93 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
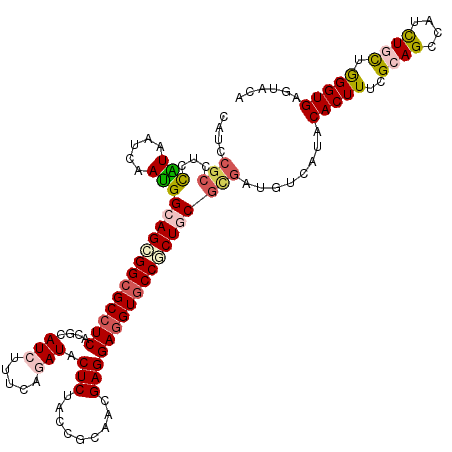
>3L_DroMel_CAF1 1268531 120 + 23771897 CAUCCGCCUCCGUUAUCAACGGCAGCGGCGCCUCGCGCAUCUUCCAGAUACUCUACCGCAACGAGGAGGUUCCGCUGCGCGACAUCAUCCACUUCAGGAGCCAUCUGCUAGGUGAGUGCA ((..(((((.(((.....)))(((((((.(((((....(((.....))).(((.........)))))))).)))))))(((......(((......)))......))).)))))..)).. ( -39.30) >DroVir_CAF1 25952 120 + 1 CAUCCGCAUCCAUAAUCAAUGGCAGCGGCGCCUCGCGUAUAUUCCAGAUACUCUAUCGCAACGAGGAGGUGCCGCUGCGCGAUGUCAUACACUUUCGCAGUCAUCUGUUGGGUGAGUAGA ((((.((...(((.....)))(((((((((((((.(((.......((.....))......)))..))))))))))))))))))).....(((((..((((....)))).)))))...... ( -44.42) >DroPse_CAF1 31109 110 + 1 ----------CGUAAUCAAUGGAAGCGGCGCCUCGCGUAUCUUUCAGAUACUCUACCGCAACGAGGAGGUGCCCCUGCGCGAUGUCAUCCACUUUCGCAGCCAUUUACUGGGUGAGUGGC ----------...........(.((.((((((((..(((((.....))))).....((...))..)))))))).)).)((((.((.....))..)))).(((((((((...))))))))) ( -35.10) >DroGri_CAF1 36337 120 + 1 CAUCCGCAUCCAUUAUAAAUGGCAGUGGCGCCUCACGCAUAUUUCAGAUACUCUACCGGAACGAGGAGGUGCCACUGCGCGAUGUCAUCCACUUUCGCAGUCAUCUGCUGGGUGAGUAAA ((((.((...(((.....)))(((((((((((((.....((((...))))(((.........))))))))))))))))))))))(((((((.....((((....)))))))))))..... ( -45.60) >DroWil_CAF1 34623 120 + 1 CAUCCGCCUCUAUUAUCAAUGGUAGCGGCGCCUCACGUAUCUUUCAGAUACUCUAUCGCAACGAGGAAGUGCCCCUUCGUGAUGUCAUACACUUUCGCAGCCAUUUGCUAGGUGAGUACA ....((((.((((((....)))))).)))).((((((((((.....)))))......((((.((((........))))((((.((.....))..))))......))))...))))).... ( -31.70) >DroMoj_CAF1 27130 120 + 1 CAUCCGCCACCAUAAUCAAUGGCAGCGGCGCCUCACGCAUCUUUCAGAUACUCUAUCGCAACGAGGAGGUGCCGCUGCGCGAUGUCAUACACUUCCGCAGCCAUCUGCUGGGUGAGUAUU ((((.((...(((.....)))(((((((((((((....(((.....))).(((.........))))))))))))))))))))))......(((..(.((((.....)))).)..)))... ( -46.60) >consensus CAUCCGCCUCCAUAAUCAAUGGCAGCGGCGCCUCACGCAUCUUUCAGAUACUCUACCGCAACGAGGAGGUGCCGCUGCGCGAUGUCAUACACUUUCGCAGCCAUCUGCUGGGUGAGUACA ....(((...(((.....)))(((((((((((((....(((.....))).(((.........)))))))))))))))))))........(((((..((((....)))).)))))...... (-31.12 = -31.93 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:43 2006