| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,268,047 – 1,268,156 |
| Length | 109 |
| Max. P | 0.709612 |

| Location | 1,268,047 – 1,268,156 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.76 |
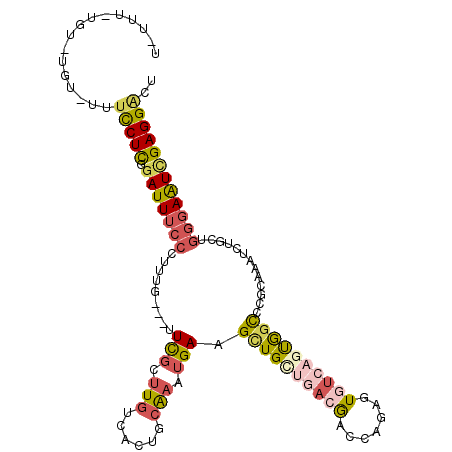
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.63 |
| Covariance contribution | 1.01 |
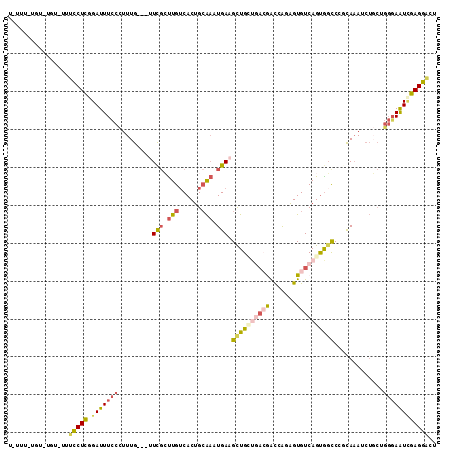
| Combinations/Pair | 1.46 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
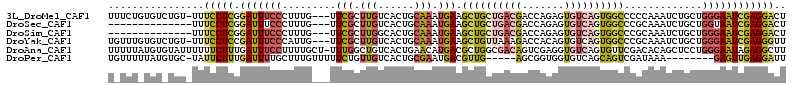
>3L_DroMel_CAF1 1268047 109 + 23771897 UUUCUGUGUCUGU-UUUCCUCGGAUUUCCCUUUG---UUCGCUUGUCACUGCAAAUGAAGCUGCUGACGACCAGAGUGUCAGUGGCCCCCAAAUCUGCUGGGAAUCGAGGACU .............-..(((((((..(((((....---((((.((((....)))).))))((..(((((.((....)))))))..)).............)))))))))))).. ( -33.90) >DroSec_CAF1 22823 96 + 1 --------------UUUCCUCGGAUUUCCCUUUG---UUCGCUUGUCACUGCAAAUGAAGCUGCUGACGACCAGAGUGUCAGUGGCCCGCAAAUCUGCUGGUAAUCGAGGACU --------------..((((((.(((.((.....---((((.((((....)))).))))((..(((((.((....)))))))..))..(((....))).)).))))))))).. ( -30.20) >DroSim_CAF1 23243 96 + 1 --------------UUUCCUCGGAUUUCCCUUUG---UUCGCUUGGCACUGCAAAUGAAGCUGCUGACGACCAGAGUGUCAGUGGCCCGCAAAUCUGCUGGGAAUCGAGGACU --------------..(((((((..(((((((..---((.((........)).))..))((..(((((.((....)))))))..))..(((....))).)))))))))))).. ( -35.40) >DroYak_CAF1 23161 109 + 1 UGUUUGUGUCUGU-UUUCCUCCGAUUUCCCAUUG---UUCGCUUGUCACUGCAAAUGAAGCUGUUAAAGACCACAGUGUCAGUGGCCCGCAAAUCUGCUGGGAAUCGAGGGUU .............-..(((((.((((.((((..(---((.((..(((((((........(((((........)))))..)))))))..)).)))....))))))))))))).. ( -29.50) >DroAna_CAF1 23394 112 + 1 UUUUUAUGUGUAUUUUUUCUUUGAUUUCCUUUUGCU-UUUGGCUGUCACUGAACAUGACGCUGGCGACAGUCGAGGUGUCAGUGUUCGACACAGCUCCUGGGAAUAGAGGCUU ...........................(((((..((-(..((((((...(((((((((((((..((.....)).))))))).))))))..))))))...)))...)))))... ( -31.10) >DroPer_CAF1 30885 99 + 1 UGUUUUUAUGUGC-UAUUCUUUGAUUUUGCUUUGUUUUUCUGUUGUCACUGCGAAUGACGUUG-----AGCGGUGGUGUCAGCAGUCGAUAAA--------GAGUUGAGGAUU .............-...((((..((....(((((((...(((((((((((((..(......).-----.)))))))...))))))..))))))--------).))..)))).. ( -26.20) >consensus U_UUU_UGU_UGU_UUUCCUCGGAUUUCCCUUUG___UUCGCUUGUCACUGCAAAUGAAGCUGCUGACGACCAGAGUGUCAGUGGCCCGCAAAUCUGCUGGGAAUCGAGGACU ................(((((.(((((((.........(((.(((......))).))).((((((((((.......)))))))))).............)))))))))))).. (-16.62 = -17.63 + 1.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:41 2006