| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,723,000 – 12,723,105 |
| Length | 105 |
| Max. P | 0.997413 |

| Location | 12,723,000 – 12,723,105 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -14.69 |
| Energy contribution | -15.01 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
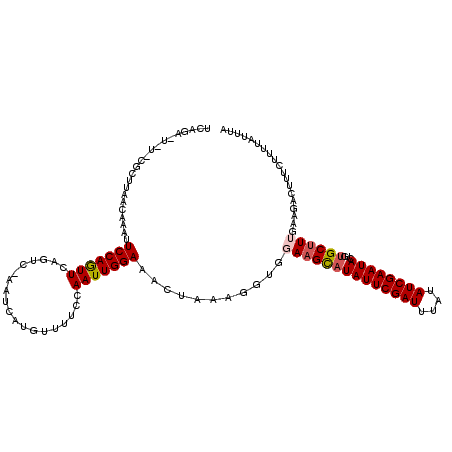
>3L_DroMel_CAF1 12723000 105 + 23771897 --------------AAACAAAUUCCAGUUCAUUC-AAUUAUGUUUUCUAAUUGGAAACUAAUGGUGGAAGCAUAUUCGAUUUAUAUCGAAUACUGUGCUUUGAAAACUUUCUUUUAUUUA --------------...........(((((((..-.((((.((((((.....)))))))))).)))((((((((((((((....)))))))...)))))))...))))............ ( -20.20) >DroSec_CAF1 3335 119 + 1 UCAGAUUAUCCGCUUAACAAAUUCCAGUUCACUG-AAUGAUGUUUUCCAAUUGGAAACUAAAGGUGGAUGCAUAUUCGAUUUAUAUCGAAUACUAUGCUUUGAAGACUUUCUUUUAUUUA (((((..((((((((((((.((((.((....)))-)))..))))((((....)))).....))))))))(((((((((((....))))))...))))))))))................. ( -28.20) >DroSim_CAF1 3710 119 + 1 UCAGAUUAUCCGCUUAACAAAUUCCAGUUCAGUC-AAUGAUGUUUUCCAAUUGGAAACUAAAGGUGGAAGCAUAUUCGAUUUAUAUCGAAUACUUUGCUUUGAAGACUUUCUUUUAUUUA (((((...(((((((......((((((((....(-(....))......)))))))).....))))))).(((((((((((....))))))))...))))))))................. ( -25.10) >DroEre_CAF1 3318 119 + 1 UCAGAAUCUACGCUCAACAAAAUCCAAUUGAGUACGAACA-AGUGUCCAAUUGGAAAAUACGGAUAAAAGUAUAUUCGAUUUAUAUCGAAUACGCUGCUUGUAAGCCUUUCCUUUAUUUA ......................((((((((..(((.....-.)))..))))))))(((((.(((....((..((((((((....))))))))..))(((....)))...)))..))))). ( -28.80) >DroYak_CAF1 3167 108 + 1 UCAGA------------UAAAAUCCAAUUGAGUUUGAACUCAACUUCCAAUUGGAAAAUAAGGAUGAAAGUAUAUUCGAUUUAUAUCGAAUACGGCGCUUGCAAGCCUUUCUUUUAUUUA ..(((------------((((((((..((((((....)))))).((((....)))).....))))((((...((((((((....)))))))).(((........))))))).))))))). ( -28.10) >consensus UCAGA_U_U_CGCUUAACAAAUUCCAGUUCAGUC_AAUCAUGUUUUCCAAUUGGAAACUAAAGGUGGAAGCAUAUUCGAUUUAUAUCGAAUACUGUGCUUUGAAGACUUUCUUUUAUUUA ......................(((((((...................)))))))...........((((((((((((((....))))))))...))))))................... (-14.69 = -15.01 + 0.32)


| Location | 12,723,000 – 12,723,105 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -11.42 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12723000 105 - 23771897 UAAAUAAAAGAAAGUUUUCAAAGCACAGUAUUCGAUAUAAAUCGAAUAUGCUUCCACCAUUAGUUUCCAAUUAGAAAACAUAAUU-GAAUGAACUGGAAUUUGUUU-------------- .(((((((............(((((...((((((((....)))))))))))))......(((((((.((((((.......)))))-)...)))))))..)))))))-------------- ( -19.10) >DroSec_CAF1 3335 119 - 1 UAAAUAAAAGAAAGUCUUCAAAGCAUAGUAUUCGAUAUAAAUCGAAUAUGCAUCCACCUUUAGUUUCCAAUUGGAAAACAUCAUU-CAGUGAACUGGAAUUUGUUAAGCGGAUAAUCUGA ........(((...........(((((...((((((....)))))))))))((((.........((((....))))((((...((-(((....)))))...))))....))))..))).. ( -26.50) >DroSim_CAF1 3710 119 - 1 UAAAUAAAAGAAAGUCUUCAAAGCAAAGUAUUCGAUAUAAAUCGAAUAUGCUUCCACCUUUAGUUUCCAAUUGGAAAACAUCAUU-GACUGAACUGGAAUUUGUUAAGCGGAUAAUCUGA ........(((..((((.(..(((((((((((((((....)))))))))..(((((..((((((((((....))))..((....)-))))))).)))))))))))..).))))..))).. ( -26.70) >DroEre_CAF1 3318 119 - 1 UAAAUAAAGGAAAGGCUUACAAGCAGCGUAUUCGAUAUAAAUCGAAUAUACUUUUAUCCGUAUUUUCCAAUUGGACACU-UGUUCGUACUCAAUUGGAUUUUGUUGAGCGUAGAUUCUGA ........(((((.(((((((((.((..((((((((....))))))))..)).............(((((((((..((.-.....))..))))))))).)))).))))).)...)))).. ( -27.30) >DroYak_CAF1 3167 108 - 1 UAAAUAAAAGAAAGGCUUGCAAGCGCCGUAUUCGAUAUAAAUCGAAUAUACUUUCAUCCUUAUUUUCCAAUUGGAAGUUGAGUUCAAACUCAAUUGGAUUUUA------------UCUGA ...((((((((((((((....)))...(((((((((....))))))))).))))).(((......(((....)))((((((((....))))))))))))))))------------).... ( -28.70) >consensus UAAAUAAAAGAAAGUCUUCAAAGCACAGUAUUCGAUAUAAAUCGAAUAUGCUUCCACCCUUAGUUUCCAAUUGGAAAACAUCAUU_GACUGAACUGGAAUUUGUUAAGCG_A_A_UCUGA ..((((((...................(((((((((....)))))))))..............(((((....)))))......................))))))............... (-11.42 = -11.90 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:45 2006