
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,722,403 – 12,722,638 |
| Length | 235 |
| Max. P | 0.908420 |

| Location | 12,722,403 – 12,722,523 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -20.15 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12722403 120 - 23771897 GUUUCGAUUAUGAACGGAAUCGCCGAGGUGCAGAAGGACAAUGAAAAGUCGGACAUGCUGCGAAAGAACAUCGAGAACAUCGAACUGGGCCUCAGCGAGGCCCAGGUGAUGAUGGCCCUG .(((((((......(((.....)))...(((((...(((........))).......))))).......))))))).(((((..(((((((((...)))))))))....)))))...... ( -38.10) >DroVir_CAF1 2876 120 - 1 GUUUCCAUAAUGAACGGUAUAUCAGAGGUGCAAAAAGAUAAUGAAAAAUCUGAUAUAUUACGAAAGAAUAUUGAAAACAUUGAACUUGGCUUGAGUGAAGCACAAGUCAUGAUGGCCCUA ....((((.((((..(((((((((((....((.........)).....))))))))))).(....)..................((((((((.....)))).)))))))).))))..... ( -25.60) >DroGri_CAF1 3332 120 - 1 GUUUCCAUCAUAAACGGUAUAUCUGAGGUUCAGAAGGACAAUGAAAAAUCGGAUAUAUUGCGAAAGAAUAUUGAAAAUAUUGAGCUUGGCCUGAGUGAGGCCCAAGUGAUGAUGGCACUU ....(((((((...((((((((((((..((((.........))))...))))))))))))(....).................(((((((((.....))).)))))).)))))))..... ( -36.50) >DroWil_CAF1 1565 120 - 1 AUUUCGAUUAUGAACGGCAUAGCUGAAGUUCAAAAGGACAAUGAGAAAUCGGAUAUGCUGAAAAAGAAUAUUGAAAAUAUUGAGCUUGGUCUGAGUGAAGCACAAGUUAUGAUGGCUCUA .(((((((......(((((((.((...((((....))))...((....)))).))))))).........))))))).....(((((..((((((.(........).))).)))))))).. ( -24.86) >DroMoj_CAF1 1715 120 - 1 GUUUCGAUUAUGAACGGAAUAUCCGAAGUUCAAAAAGAUAAUGAGAAAUCUGAUAUAUUACGAAAGAACAUAGAAAGCAUUGAACUUGGCUUGAGCGAGGCGCAGGUCAUGAUGGCCCUA ((((((.(((....(((.....)))((((((((...((((((.((....)).)).)))).(....).............))))))))....))).))))))...((((.....))))... ( -26.00) >DroAna_CAF1 2995 120 - 1 GUCUCGAUUAUGAACGGCAUUUCUGAGGUGCAGAAGGACAACGAGAAGUCGGACAUGCUGCGGAAGAACAUUGAAAACAUCGAACUGGGUCUGAGCGAGGCCCAGGUGAUGAUGGCCCUC ((((((((......(((((((((((.....))))))(((........))).....)))))(....)...)))))...(((((..((((((((.....)))))))).)))))..))).... ( -34.60) >consensus GUUUCGAUUAUGAACGGAAUAUCUGAGGUGCAAAAGGACAAUGAAAAAUCGGAUAUACUGCGAAAGAACAUUGAAAACAUUGAACUUGGCCUGAGCGAGGCCCAAGUGAUGAUGGCCCUA .((((.........((((((((((((......................))))))))))))(....)......)))).((((..(((((((((.....)))).)))))...))))...... (-20.15 = -20.18 + 0.03)

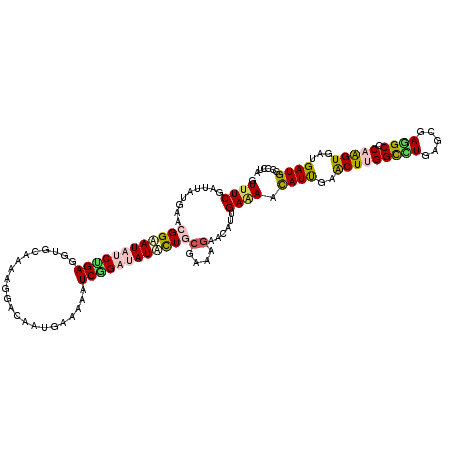

| Location | 12,722,443 – 12,722,563 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -15.98 |
| Energy contribution | -14.48 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12722443 120 + 23771897 AUGUUCUCGAUGUUCUUUCGCAGCAUGUCCGACUUUUCAUUGUCCUUCUGCACCUCGGCGAUUCCGUUCAUAAUCGAAACAUGCUCCACUCGCAAGGCUUCGAGGCCUUGAAGGACUGUU ...........((((((..(.(((((((..(((........)))..............(((((........)))))..))))))).).....(((((((....))))))))))))).... ( -28.60) >DroVir_CAF1 2916 120 + 1 AUGUUUUCAAUAUUCUUUCGUAAUAUAUCAGAUUUUUCAUUAUCUUUUUGCACCUCUGAUAUACCGUUCAUUAUGGAAACAUGUUCGACCCGCAAGGCUUCCAAGCCUUGCAAAACAGUU .((((((................(((((((((.....((.........))....)))))))))..(((...((((....))))...)))..((((((((....))))))))))))))... ( -29.70) >DroGri_CAF1 3372 120 + 1 AUAUUUUCAAUAUUCUUUCGCAAUAUAUCCGAUUUUUCAUUGUCCUUCUGAACCUCAGAUAUACCGUUUAUGAUGGAAACAUGUUCAACGCGCAAAGCUUCCAAGCCCUGCAACACAGUU ...................((..((((((.((...((((.........))))..)).))))))..(((.(..(((....)))..).)))))(((..(((....)))..)))......... ( -18.20) >DroWil_CAF1 1605 120 + 1 AUAUUUUCAAUAUUCUUUUUCAGCAUAUCCGAUUUCUCAUUGUCCUUUUGAACUUCAGCUAUGCCGUUCAUAAUCGAAAUAUGUUCGACACGCAGGGCCUCCAGGCCCUGCAGAACUGUU ......................(((((.(.((....(((.........)))...)).).))))).((((....(((((.....)))))...((((((((....)))))))).)))).... ( -32.00) >DroMoj_CAF1 1755 120 + 1 AUGCUUUCUAUGUUCUUUCGUAAUAUAUCAGAUUUCUCAUUAUCUUUUUGAACUUCGGAUAUUCCGUUCAUAAUCGAAACAUGUUCAACGCGCAAGGCUUCCAAGCCUUGCAAAACAGUU ..((....((((......)))).......((((........))))..((((((..(((.....)))(((......)))....)))))).))((((((((....))))))))......... ( -25.00) >DroAna_CAF1 3035 120 + 1 AUGUUUUCAAUGUUCUUCCGCAGCAUGUCCGACUUCUCGUUGUCCUUCUGCACCUCAGAAAUGCCGUUCAUAAUCGAGACAUGCUCCACUCGCAGGGCUUCCAGGCCCUGGAGCACUGUC .......((.(((((....(.(((((((((((.........((..(((((.....)))))..)).........))).)))))))).).....(((((((....)))))))))))).)).. ( -37.47) >consensus AUGUUUUCAAUAUUCUUUCGCAACAUAUCCGAUUUCUCAUUGUCCUUCUGAACCUCAGAUAUACCGUUCAUAAUCGAAACAUGUUCAACGCGCAAGGCUUCCAAGCCCUGCAAAACAGUU .....................(((((....(((........)))......................(((......)))..)))))......((((((((....))))))))......... (-15.98 = -14.48 + -1.50)


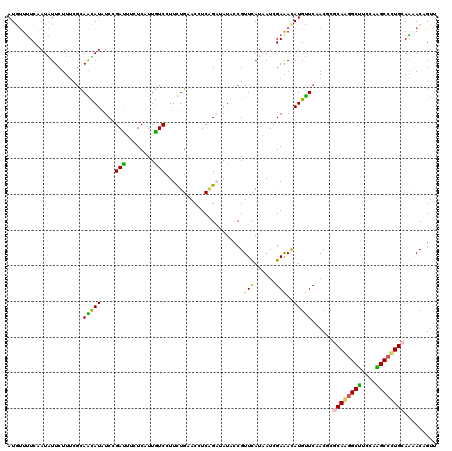
| Location | 12,722,443 – 12,722,563 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -34.29 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12722443 120 - 23771897 AACAGUCCUUCAAGGCCUCGAAGCCUUGCGAGUGGAGCAUGUUUCGAUUAUGAACGGAAUCGCCGAGGUGCAGAAGGACAAUGAAAAGUCGGACAUGCUGCGAAAGAACAUCGAGAACAU ....(((((((...((((((((((..(((.......))).)))))((((........))))...)))))...))))))).........(((...(((...(....)..))))))...... ( -32.90) >DroVir_CAF1 2916 120 - 1 AACUGUUUUGCAAGGCUUGGAAGCCUUGCGGGUCGAACAUGUUUCCAUAAUGAACGGUAUAUCAGAGGUGCAAAAAGAUAAUGAAAAAUCUGAUAUAUUACGAAAGAAUAUUGAAAACAU ...((.(((((((((((....))))))))))).))...((((((.((........(((((((((((....((.........)).....))))))))))).(....).....)).)))))) ( -30.80) >DroGri_CAF1 3372 120 - 1 AACUGUGUUGCAGGGCUUGGAAGCUUUGCGCGUUGAACAUGUUUCCAUCAUAAACGGUAUAUCUGAGGUUCAGAAGGACAAUGAAAAAUCGGAUAUAUUGCGAAAGAAUAUUGAAAAUAU ..(..(((.((((((((....)))))))))))..)...((((((.((.......((((((((((((..((((.........))))...))))))))))))(....).....)).)))))) ( -30.00) >DroWil_CAF1 1605 120 - 1 AACAGUUCUGCAGGGCCUGGAGGCCCUGCGUGUCGAACAUAUUUCGAUUAUGAACGGCAUAGCUGAAGUUCAAAAGGACAAUGAGAAAUCGGAUAUGCUGAAAAAGAAUAUUGAAAAUAU ....(((((((((((((....))))))))..((((((.....))))))......(((((((.((...((((....))))...((....)))).)))))))....)))))........... ( -39.70) >DroMoj_CAF1 1755 120 - 1 AACUGUUUUGCAAGGCUUGGAAGCCUUGCGCGUUGAACAUGUUUCGAUUAUGAACGGAAUAUCCGAAGUUCAAAAAGAUAAUGAGAAAUCUGAUAUAUUACGAAAGAACAUAGAAAGCAU ...((((((((((((((....))))))))...((((((((((((((........)))))))).....))))))...((((((.((....)).)).)))).(....).......)))))). ( -28.30) >DroAna_CAF1 3035 120 - 1 GACAGUGCUCCAGGGCCUGGAAGCCCUGCGAGUGGAGCAUGUCUCGAUUAUGAACGGCAUUUCUGAGGUGCAGAAGGACAACGAGAAGUCGGACAUGCUGCGGAAGAACAUUGAAAACAU ..((((((((((((((......)))))).)))((.((((((((.(((((.......(..((((((.....))))))..).......))))))))))))).))......)))))....... ( -44.04) >consensus AACAGUUCUGCAAGGCCUGGAAGCCUUGCGAGUCGAACAUGUUUCGAUUAUGAACGGAAUAUCUGAGGUGCAAAAGGACAAUGAAAAAUCGGAUAUACUGCGAAAGAACAUUGAAAACAU ........((((((((......))))))))........(((((((.........((((((((((((......................))))))))))))(....)......))).)))) (-21.71 = -21.88 + 0.17)

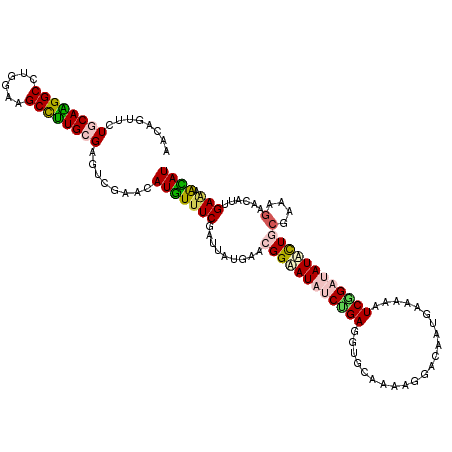
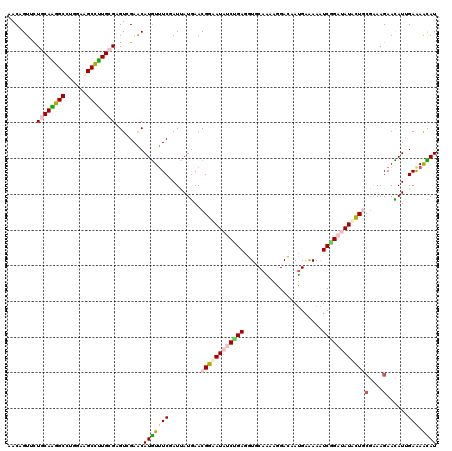
| Location | 12,722,523 – 12,722,638 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.64 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.37 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12722523 115 - 23771897 -----GUUUUGUAGACAGAAAAUUGAGAAAAUCGGCAAAAUGACGCAAAUGUCGCAGGACGAAAUCAUAACCAAUACGAAAACAGUCCUUCAAGGCCUCGAAGCCUUGCGAGUGGAGCAU -----(((((((.((((......(((.....)))((........))...)))))))))))......................((.((...((((((......)))))).)).))...... ( -23.40) >DroVir_CAF1 2996 112 - 1 --------CUGCAGACAGAAAAUUGAGCAAAUCGGCAGAAUGACGCAGAUGUCGCAAGACGAAAUUAUAAGCAAUACAAAAACUGUUUUGCAAGGCUUGGAAGCCUUGCGGGUCGAACAU --------.....(((..........((..(((.((........)).)))(((....)))..........))...............((((((((((....)))))))))))))...... ( -29.40) >DroPse_CAF1 5165 114 - 1 ------UGUUCCAGACAGAAAAUUGAAAAAAUCGGCAAAAUGACUCAAAUGUCACAGGACGAAAUAAUAACCAAUACAAAAACAGUAUUGCAAGGCCUAGAGGCUUUGCGAGUGGAGCAU ------(((((((..................(((.(....((((......))))...).))).........................((((((((((....)))))))))).))))))). ( -30.20) >DroGri_CAF1 3452 109 - 1 -----------CAGACAGAAAAUAGAGCAAAUCGGCAAAAUGACGCAAAUGUCGCAAGACGAAAUAAUAAGCAAUACAAAAACUGUGUUGCAGGGCUUGGAAGCUUUGCGCGUUGAACAU -----------...............((......)).....(((((...((((....)))).........((((((((.....))))))))((((((....))))))..)))))...... ( -28.00) >DroMoj_CAF1 1835 120 - 1 UUCUUUUUCUGCAGACAGAAAAUUGAGCAAAUCGGCAAAAUGACGCAGAUGUCGCAAGACGAAAUUAUAAGCAAUACAAAAACUGUUUUGCAAGGCUUGGAAGCCUUGCGCGUUGAACAU .((.(((((((....)))))))..))((......)).....(((((...((((....))))............................((((((((....)))))))))))))...... ( -34.00) >DroAna_CAF1 3115 115 - 1 -----UAUUUGCAGACAGAAAAUCGAAAAAAUCGGAAAAAUGACGCAAAUGUCGCAGGACGAAAUAAUAUCCAAUACGAAGACAGUGCUCCAGGGCCUGGAAGCCCUGCGAGUGGAGCAU -----.((((((.(.((.....((((.....)))).....)).)))))))(((....))).........(((...((.......))((((((((((......)))))).))))))).... ( -28.20) >consensus ______UUUUGCAGACAGAAAAUUGAGAAAAUCGGCAAAAUGACGCAAAUGUCGCAAGACGAAAUAAUAACCAAUACAAAAACAGUGUUGCAAGGCCUGGAAGCCUUGCGAGUGGAACAU ...............................((.((.............((((....))))...........................((((((((......)))))))).)).)).... (-17.45 = -17.37 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:43 2006