| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,709,118 – 12,709,208 |
| Length | 90 |
| Max. P | 0.941683 |

| Location | 12,709,118 – 12,709,208 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -20.75 |
| Energy contribution | -21.87 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
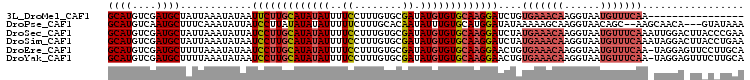
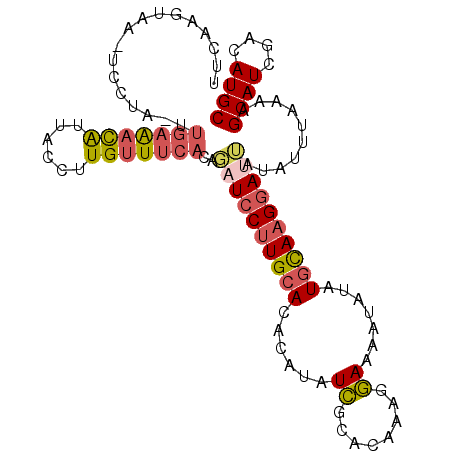
>3L_DroMel_CAF1 12709118 90 + 23771897 GCAUGUCGAUGCUAUUAAAUAUAAUUCUUGCAUAUAUUUUCCUUUGUGCGAUAUGUGUGCAAGGAUCUGUGAAACAAGGUAAUGUUUCAA---------------- ((((....))))...........((((((((((((((..(((.....).)).))))))))))))))...(((((((......))))))).---------------- ( -24.80) >DroPse_CAF1 554 101 + 1 GCAUGUCAAUGCUUUCAAAUAUUAUCCUUAUAUAUAUUUUUCUUUGCACAAUAUUUGUGCAUGGAUAUAAAAAGCAAGGUAACAGC--AAGCAACA---GUAUAAA ((.(((...(((((((......(((((.................((((((.....)))))).)))))......).))))))...))--).))....---....... ( -17.47) >DroSec_CAF1 1543 106 + 1 GCAUGUCGAUGCUAUUAAAUAUUAUCCUUGCAUAUAUUUUCCUUUGUGCGAUAUGUGUGCAAGGAUCUAUGAAACAAGGUAAUGUUUCAAAUUGGACUUACCCGAA ((((....))))...........((((((((((((((..(((.....).)).))))))))))))))...(((((((......)))))))..((((......)))). ( -30.40) >DroSim_CAF1 1518 106 + 1 GCAUGUCGAUGCUAUUAAAUAUAAUCCUUGCAUAUAUUUUCCUUUGUGCGAUAUGUGUGCAAGGAUCUAUGAAACAAGGUAAUGUUUCAAAUAGGACUUACCUGAA ((((....)))).......((((((((((((((((((..(((.....).)).)))))))))))))).)))).....((((((.((((......))))))))))... ( -30.30) >DroEre_CAF1 1490 105 + 1 GCAUGUCGAUGCUUUUAAAUAUAAUCCUUGCAUAUAUUUUCCUUUGUGCGAUAUGUGUGCAAGGAACUGUGAAACAAGGUAAUGUUUCAA-UAGGAGUUCCUUGCA ((((((((.(((....(((((((.........)))))))......))))))))))).(((((((((((.(((((((......))))))).-....))))))))))) ( -34.10) >DroYak_CAF1 1527 105 + 1 GCAUGUCGAUGCUUUUAAAUAUAAUCCUUGCAUAUAUUUUCCUUUGUGCGAUAUGUGUGCAAGGAACUGUGAAACAAGGUAAUGUUUCAA-UAGGAGUUUCUUGCA (((....((.(((((((.......(((((((((((((..(((.....).)).)))))))))))))....(((((((......))))))).-))))))).)).))). ( -33.30) >consensus GCAUGUCGAUGCUAUUAAAUAUAAUCCUUGCAUAUAUUUUCCUUUGUGCGAUAUGUGUGCAAGGAUCUAUGAAACAAGGUAAUGUUUCAA_UAGGA_UUACCUGAA ((((....))))............(((((((((((((..((........)).)))))))))))))....(((((((......)))))))................. (-20.75 = -21.87 + 1.11)
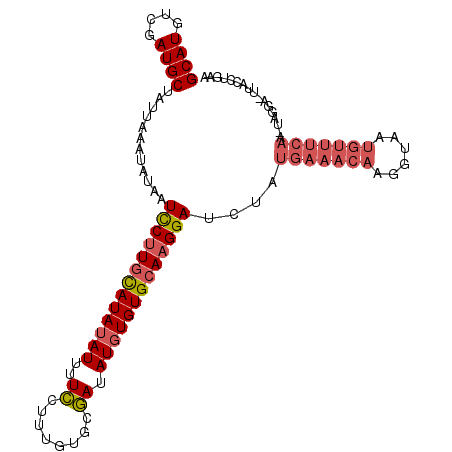
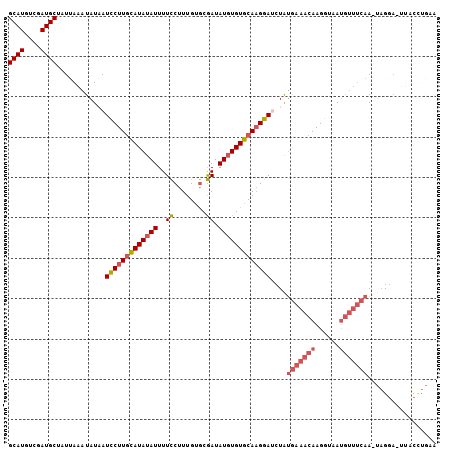
| Location | 12,709,118 – 12,709,208 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -14.09 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12709118 90 - 23771897 ----------------UUGAAACAUUACCUUGUUUCACAGAUCCUUGCACACAUAUCGCACAAAGGAAAAUAUAUGCAAGAAUUAUAUUUAAUAGCAUCGACAUGC ----------------.(((((((......)))))))....(((((((.........)))...))))(((((((.........)))))))....((((....)))) ( -14.40) >DroPse_CAF1 554 101 - 1 UUUAUAC---UGUUGCUU--GCUGUUACCUUGCUUUUUAUAUCCAUGCACAAAUAUUGUGCAAAGAAAAAUAUAUAUAAGGAUAAUAUUUGAAAGCAUUGACAUGC .......---........--(((((((...(((((((..(((((.((((((.....))))))........((....)).)))))......))))))).))))).)) ( -19.80) >DroSec_CAF1 1543 106 - 1 UUCGGGUAAGUCCAAUUUGAAACAUUACCUUGUUUCAUAGAUCCUUGCACACAUAUCGCACAAAGGAAAAUAUAUGCAAGGAUAAUAUUUAAUAGCAUCGACAUGC .....(((.(((.....(((((((......)))))))...(((((((((......((........)).......)))))))))................))).))) ( -21.82) >DroSim_CAF1 1518 106 - 1 UUCAGGUAAGUCCUAUUUGAAACAUUACCUUGUUUCAUAGAUCCUUGCACACAUAUCGCACAAAGGAAAAUAUAUGCAAGGAUUAUAUUUAAUAGCAUCGACAUGC .....(((.(((((((((((((((......)))))))..((((((((((......((........)).......))))))))))......)))))....))).))) ( -23.52) >DroEre_CAF1 1490 105 - 1 UGCAAGGAACUCCUA-UUGAAACAUUACCUUGUUUCACAGUUCCUUGCACACAUAUCGCACAAAGGAAAAUAUAUGCAAGGAUUAUAUUUAAAAGCAUCGACAUGC (((((((((((....-.(((((((......))))))).)))))))))))..(((.(((.........(((((((.........)))))))........))).))). ( -26.63) >DroYak_CAF1 1527 105 - 1 UGCAAGAAACUCCUA-UUGAAACAUUACCUUGUUUCACAGUUCCUUGCACACAUAUCGCACAAAGGAAAAUAUAUGCAAGGAUUAUAUUUAAAAGCAUCGACAUGC ((((((.((((....-.(((((((......))))))).)))).))))))..(((.(((.........(((((((.........)))))))........))).))). ( -19.83) >consensus UUCAAGUAA_UCCUA_UUGAAACAUUACCUUGUUUCACAGAUCCUUGCACACAUAUCGCACAAAGGAAAAUAUAUGCAAGGAUUAUAUUUAAAAGCAUCGACAUGC .................(((((((......)))))))..((((((((((......((........)).......))))))))))..........((((....)))) (-14.09 = -14.70 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:39 2006