
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,706,872 – 12,706,983 |
| Length | 111 |
| Max. P | 0.765144 |

| Location | 12,706,872 – 12,706,983 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
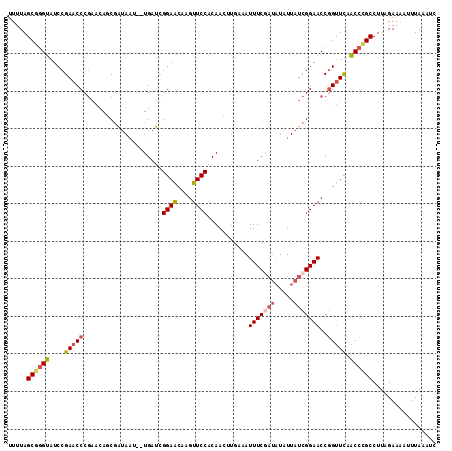
>3L_DroMel_CAF1 12706872 111 + 23771897 UUUUAGCGGGUAUCCGAACCCGAAUAGCGAUAAU--UGAUCGGAGCAAGUUCCACAAUUUGACAUUUCGAUAUAUUAUCGGAACCGGUUCAACCUGCCUUUGAAAACUUAAA-C .....((((((....(((((.(.....(((((((--..((((((.((((((....))))))....))))))..)))))))...).))))).))))))...............-. ( -27.20) >DroSec_CAF1 70174 112 + 1 UUUUAGCGGGUAUCCGAACCCGAACAGCGAUAAU--UGAUGGGAACAAGUUCCACAACUUGAAAUUUCGAUAUAUUAUCGGAACCGGUUCCACCCGCCUUAGAAAAUUUAAAUC (((((((((((....(((((.(.....(((((((--..((.((((((((((....))))))...)))).))..)))))))...).))))).))))))..))))).......... ( -28.10) >DroSim_CAF1 68996 112 + 1 UUUUAGCGGGUAUCCGAACCCAAACAGCGAUAAU--UGAUCGGAACAAGUUCCACAACUAGAAAUUUCGAUAUAUUAUCGGAACCGGUUCAACCCGCCUUUGAAAAUUUAAGUC .....((((((....(((((.......(((((((--..(((((((....(((........))).)))))))..))))))).....))))).))))))................. ( -27.90) >DroEre_CAF1 73538 114 + 1 AUUUAGCAGGUAUCCGAACCCGUGCACCCGAACCCGUGACCGGAACCAGUUCCACAAGAUUUUAUUUCACUAUUUUAUCGGAACCUGCUUAGCCCGCCUUCGAACAUUUAAAUU ....(((((((.(((((..(((..(((........)))..)))..............((.......)).........))))))))))))......................... ( -24.90) >DroYak_CAF1 73387 112 + 1 UCUUAGCGAGUAUCCGAACCCGAGUACCGAUAAC--UGAUCGGAACCAGUUCCACAAGCUUUCAUUUCAACCUAUUAUCGGAACCGGUUCAGCCCGCCUUAGAACAUUUAAUUU (((..(((.(((..((....))..)))((((...--..))))(((((.(((((..........................))))).)))))....)))...)))........... ( -23.47) >consensus UUUUAGCGGGUAUCCGAACCCGAACAGCGAUAAU__UGAUCGGAACAAGUUCCACAACUUGAAAUUUCGAUAUAUUAUCGGAACCGGUUCAACCCGCCUUAGAAAAUUUAAAUC .....((((((....(((((.....................((((....))))...........(((((((.....)))))))..))))).))))))................. (-17.96 = -18.88 + 0.92)

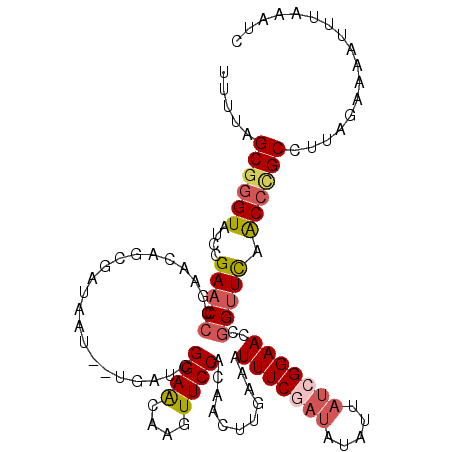
| Location | 12,706,872 – 12,706,983 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.92 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12706872 111 - 23771897 G-UUUAAGUUUUCAAAGGCAGGUUGAACCGGUUCCGAUAAUAUAUCGAAAUGUCAAAUUGUGGAACUUGCUCCGAUCA--AUUAUCGCUAUUCGGGUUCGGAUACCCGCUAAAA .-..............(((.(((((((((.(...(((((((..(((((....)).......(((......))))))..--))))))).....).))))))...))).))).... ( -24.50) >DroSec_CAF1 70174 112 - 1 GAUUUAAAUUUUCUAAGGCGGGUGGAACCGGUUCCGAUAAUAUAUCGAAAUUUCAAGUUGUGGAACUUGUUCCCAUCA--AUUAUCGCUGUUCGGGUUCGGAUACCCGCUAAAA ................((((((((...((((..((((.........((....)).(((((.((((....))))...))--)))........))))..)))).)))))))).... ( -31.90) >DroSim_CAF1 68996 112 - 1 GACUUAAAUUUUCAAAGGCGGGUUGAACCGGUUCCGAUAAUAUAUCGAAAUUUCUAGUUGUGGAACUUGUUCCGAUCA--AUUAUCGCUGUUUGGGUUCGGAUACCCGCUAAAA ................(((((((((((((.(..((((((((..((((.(((....((((....)))).))).))))..--)))))))..)..).))))))...))))))).... ( -31.60) >DroEre_CAF1 73538 114 - 1 AAUUUAAAUGUUCGAAGGCGGGCUAAGCAGGUUCCGAUAAAAUAGUGAAAUAAAAUCUUGUGGAACUGGUUCCGGUCACGGGUUCGGGUGCACGGGUUCGGAUACCUGCUAAAU .........(((((....)))))..((((((((((((...................((((((.(.((((..(((....)))..)))).).)))))).))))).))))))).... ( -37.15) >DroYak_CAF1 73387 112 - 1 AAAUUAAAUGUUCUAAGGCGGGCUGAACCGGUUCCGAUAAUAGGUUGAAAUGAAAGCUUGUGGAACUGGUUCCGAUCA--GUUAUCGGUACUCGGGUUCGGAUACUCGCUAAGA ................((((((((((((((((.(((((.(((((((........)))))))..(((((((....))))--)))))))).)))..)))))))...)))))).... ( -36.20) >consensus GAUUUAAAUUUUCAAAGGCGGGUUGAACCGGUUCCGAUAAUAUAUCGAAAUGUCAACUUGUGGAACUUGUUCCGAUCA__AUUAUCGCUGUUCGGGUUCGGAUACCCGCUAAAA ................(((.((((((((.((((((.(((((...............))))))))))).))).)))))................((((......))))))).... (-22.00 = -21.92 + -0.08)

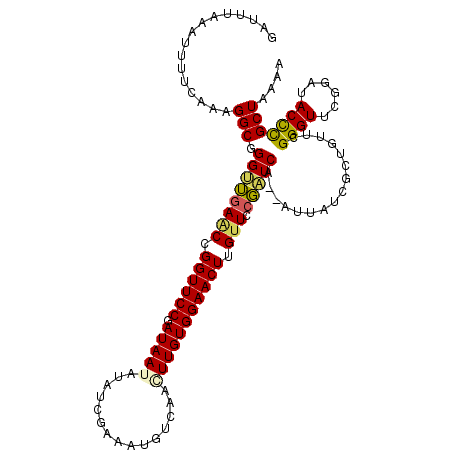

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:37 2006