
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,704,641 – 12,704,923 |
| Length | 282 |
| Max. P | 0.999818 |

| Location | 12,704,641 – 12,704,735 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -14.68 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12704641 94 - 23771897 AUCUGCUUAAGAUAGGAGAGAAAAUAAAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAACAAAUGUUUGCACUU .(((.((......)).)))....................(((......))).......((((((.....))))))(((((....)))))..... ( -14.60) >DroSec_CAF1 68283 94 - 1 AUCUGCUUAAGAUAGGAGAGAAAAUGAAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAACAAAUGUUUGCACUU .(((.((......)).)))....................(((......))).......((((((.....))))))(((((....)))))..... ( -14.60) >DroSim_CAF1 67036 94 - 1 AUCUGCUUAAGAUAGGAGAGAAAAUGAAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAACAAAUGUUUGCACUU .(((.((......)).)))....................(((......))).......((((((.....))))))(((((....)))))..... ( -14.60) >DroEre_CAF1 70377 87 - 1 AUCUGCUUAAGAUAGGAGA-------AAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAACAAAUGUUUGCACUU ((((.....)))).(((..-------........)))..(((......))).......((((((.....))))))(((((....)))))..... ( -14.80) >DroYak_CAF1 70957 87 - 1 AUCUGCUUAAGAUAGGAGA-------AAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAACAAAUGUUUGCACUU ((((.....)))).(((..-------........)))..(((......))).......((((((.....))))))(((((....)))))..... ( -14.80) >consensus AUCUGCUUAAGAUAGGAGAGAAAAU_AAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAACAAAUGUUUGCACUU .(((.((......)).)))....................(((......))).......((((((.....))))))(((((....)))))..... (-14.28 = -14.28 + 0.00)



| Location | 12,704,655 – 12,704,764 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -15.18 |
| Consensus MFE | -14.83 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12704655 109 - 23771897 AAACGAAUCGCAAAAUUAACAAGAUAAGCAUCUGCUUAAGAUAGGAGAGAAAAUAAAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAAC .........((((((((((.....(((((....))))).....(((.................)))..(((......)))..))))))))).)................ ( -14.83) >DroSec_CAF1 68297 109 - 1 AAACGAAUCGCAAAAUUAACAAGAUAAGCAUCUGCUUAAGAUAGGAGAGAAAAUGAAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAAC .........((((((((((.....(((((....))))).....(((.................)))..(((......)))..))))))))).)................ ( -14.83) >DroSim_CAF1 67050 109 - 1 AAACGAAUCGCAAAAUUAACAAGAUAAGCAUCUGCUUAAGAUAGGAGAGAAAAUGAAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAAC .........((((((((((.....(((((....))))).....(((.................)))..(((......)))..))))))))).)................ ( -14.83) >DroEre_CAF1 70391 102 - 1 AAAUGAAUCGCAAAAUUAACAAGAUAAGCAUCUGCUUAAGAUAGGAGA-------AAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAAC ...(((....(((((((((.....(((((....))))).....(((..-------........)))..(((......)))..)))))))))........)))....... ( -15.70) >DroYak_CAF1 70971 102 - 1 AAAUGAAUCGCAAAAUUAACAAGAUAAGCAUCUGCUUAAGAUAGGAGA-------AAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAAC ...(((....(((((((((.....(((((....))))).....(((..-------........)))..(((......)))..)))))))))........)))....... ( -15.70) >consensus AAACGAAUCGCAAAAUUAACAAGAUAAGCAUCUGCUUAAGAUAGGAGAGAAAAU_AAAUAAAAUCCACUGGGUACAACCAAAUUAAUUUUGACAUAAUGUCAGACAAAC .........((((((((((.....(((((....))))).....(((.................)))..(((......)))..))))))))).)................ (-14.83 = -14.83 + 0.00)

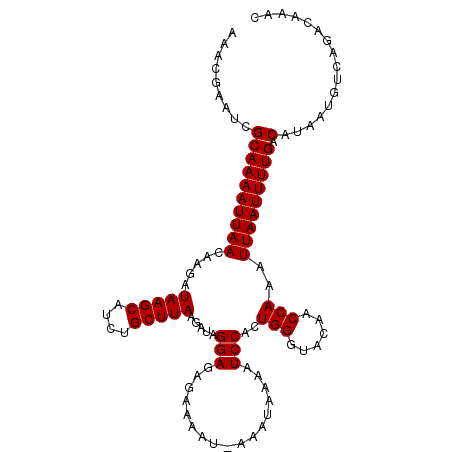
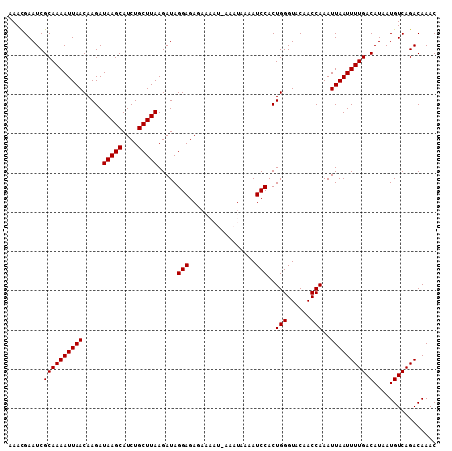
| Location | 12,704,735 – 12,704,843 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 97.79 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -21.19 |
| Energy contribution | -20.79 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12704735 108 - 23771897 AAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGU-UUGCCAUAAAACGAAUCGCAAAAUUAACAAGAUAAGC .(((...(((((..((.(((((((..........................)))))))))...))))).(((-(((........)))))).............))).... ( -24.57) >DroSec_CAF1 68377 108 - 1 AAUCGAAAAUGCAUGCCGCGGCCAGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGU-UUGCCAUAAAACGAAUCGCAAAAUUAACAAGAUAAGC .(((...(((((..((.((((((............................))))))))...))))).(((-(((........)))))).............))).... ( -22.99) >DroSim_CAF1 67130 108 - 1 AAUCGAAAAUGCAUGCCGCGGCCAGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGU-UUGCUAUAAAACGAAUCGCAAAAUUAACAAGAUAAGC .(((...(((((..((.((((((............................))))))))...))))).(((-(((........)))))).............))).... ( -22.99) >DroEre_CAF1 70464 108 - 1 AAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGA-UUGCCAUAAAAUGAAUCGCAAAAUUAACAAGAUAAGC .(((...(((((..((.(((((((..........................)))))))))...)))))(.((-((...........)))).)...........))).... ( -22.77) >DroYak_CAF1 71044 109 - 1 AAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGUUUUGCCAUAAAAUGAAUCGCAAAAUUAACAAGAUAAGC .(((...(((((..((.(((((((..........................)))))))))...))))).((((((((.((.......)).)))))))).....))).... ( -26.17) >consensus AAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGU_UUGCCAUAAAACGAAUCGCAAAAUUAACAAGAUAAGC ..(((..(((((..((.((((((............................))))))))...))))).((.....))......)))....................... (-21.19 = -20.79 + -0.40)

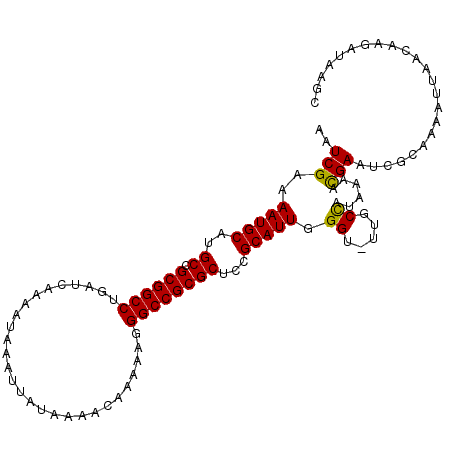

| Location | 12,704,764 – 12,704,883 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -30.69 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12704764 119 + 23771897 UAUGGCAA-ACCCAAUGCGGAGCGCGGCCCUUUUUGUUUUAUAAUUUAUUUUGAUCAGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGGAGGAACAUGUUUUGCUCAU .(((((((-(...((((((...(((((((............................)))))))...)))))).(((((....)))))..(((((((...)))))))....))))).))) ( -33.09) >DroSec_CAF1 68406 119 + 1 UAUGGCAA-ACCCAAUGCGGAGCGCGGCCCUUUUUGUUUUAUAAUUUAUUUUGAUCUGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGGAGGAACAUGUUUUGCUCAU .(((((((-(...((((((...(((((((...((.....((.....))....))...)))))))...)))))).(((((....)))))..(((((((...)))))))....))))).))) ( -32.90) >DroSim_CAF1 67159 119 + 1 UAUAGCAA-ACCCAAUGCGGAGCGCGGCCCUUUUUGUUUUAUAAUUUAUUUUGAUCUGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGGAGGAACAUGUUUUGCUCAU ...(((((-(...((((((...(((((((...((.....((.....))....))...)))))))...)))))).(((((....)))))..(((((((...)))))))....))))))... ( -32.40) >DroEre_CAF1 70493 119 + 1 UAUGGCAA-UCCCAAUGCGGAGCGCGGCCCUUUUUGUUUUAUAAUUUAUUUUGAUCAGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGCAGGAACAUGUUUUGCUCAU .(((((((-..(.((((((...(((((((............................)))))))...)))))).(((((....)))))..((((((.....))))))..)..)))).))) ( -32.39) >DroYak_CAF1 71073 120 + 1 UAUGGCAAAACCCAAUGCGGAGCGCGGCCCUUUUUGUUUUAUAAUUUAUUUUGAUCAGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGCGGAAACAUGUUUUGCUCAU .((((((((((((((((((...(((((((............................)))))))...)))))..(((((....))))))))((((......))))....))))))).))) ( -33.99) >consensus UAUGGCAA_ACCCAAUGCGGAGCGCGGCCCUUUUUGUUUUAUAAUUUAUUUUGAUCAGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGGAGGAACAUGUUUUGCUCAU ...(((((.((..((((((...(((((((............................)))))))...)))))).(((((....)))))..((((((.....))))))..)).)))))... (-30.69 = -30.77 + 0.08)

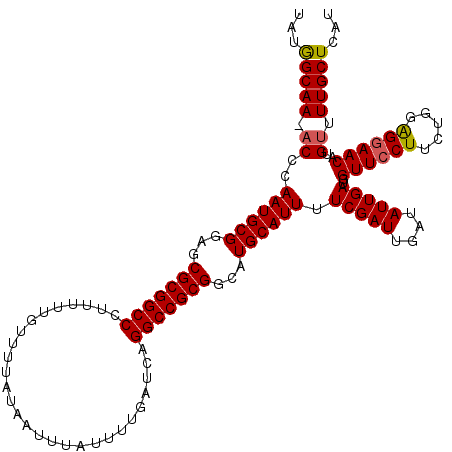
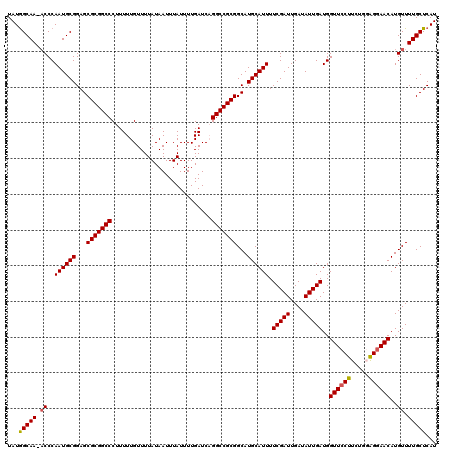
| Location | 12,704,764 – 12,704,883 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -29.65 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12704764 119 - 23771897 AUGAGCAAAACAUGUUCCUCCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGU-UUGCCAUA (((.(((((.(..((((((.....)))))).................(((((..((.(((((((..........................)))))))))...))))).).)-))))))). ( -32.07) >DroSec_CAF1 68406 119 - 1 AUGAGCAAAACAUGUUCCUCCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCAGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGU-UUGCCAUA (((.(((((.(..((((((.....)))))).................(((((..((.((((((............................))))))))...))))).).)-))))))). ( -30.49) >DroSim_CAF1 67159 119 - 1 AUGAGCAAAACAUGUUCCUCCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCAGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGU-UUGCUAUA ...((((((.(..((((((.....)))))).................(((((..((.((((((............................))))))))...))))).).)-)))))... ( -30.29) >DroEre_CAF1 70493 119 - 1 AUGAGCAAAACAUGUUCCUGCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGA-UUGCCAUA (((.((((..(..((((((.....)))))).................(((((..((.(((((((..........................)))))))))...))))).)..-))))))). ( -33.37) >DroYak_CAF1 71073 120 - 1 AUGAGCAAAACAUGUUUCCGCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGUUUUGCCAUA (((.((((((((((.((((......)))).)))..............(((((..((.(((((((..........................)))))))))...)))))..)))))))))). ( -35.37) >consensus AUGAGCAAAACAUGUUCCUCCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAUAAAACAAAAAGGGCCGCGCUCCGCAUUGGGU_UUGCCAUA (((.((((.((..((((((.....)))))).................(((((..((.((((((............................))))))))...)))))..)).))))))). (-29.65 = -29.73 + 0.08)

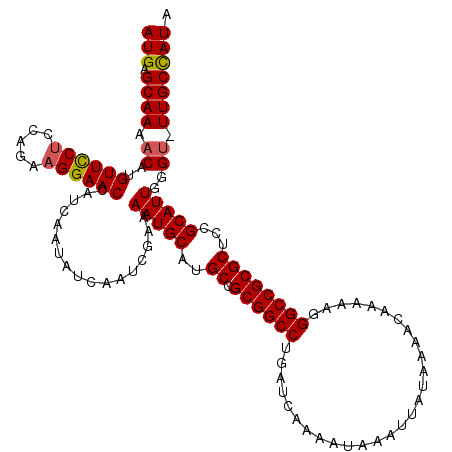

| Location | 12,704,803 – 12,704,923 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12704803 120 + 23771897 AUAAUUUAUUUUGAUCAGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGGAGGAACAUGUUUUGCUCAUCAAUGAAUUGGCGCUGAACCUGUUUGCCCUGCCUCAUUUA ............(((.((((.(.((((.(((..((((.(..((((((((((((((((...)))))))..(.....).)))))))..))..)...))))..))).))))).)))).))).. ( -30.70) >DroSec_CAF1 68445 120 + 1 AUAAUUUAUUUUGAUCUGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGGAGGAACAUGUUUUGCUCAUCAAUGAAUUGGCGCUGAACCUGUUUGCCCUGCCUCAUUUA .................(((.(.((((.(((..((((.(..((((((((((((((((...)))))))..(.....).)))))))..))..)...))))..))).))))).)))....... ( -29.30) >DroSim_CAF1 67198 120 + 1 AUAAUUUAUUUUGAUCUGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGGAGGAACAUGUUUUGCUCAUCAAUGAAUUGGCGCUGAACCUGUUUGCCCUGCCUCAUUUA .................(((.(.((((.(((..((((.(..((((((((((((((((...)))))))..(.....).)))))))..))..)...))))..))).))))).)))....... ( -29.30) >DroEre_CAF1 70532 120 + 1 AUAAUUUAUUUUGAUCAGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGCAGGAACAUGUUUUGCUCAUCAAUGAAUUGGCUCUGAACCUGUUUGCCCUGCCUCAUUUA ............(((.((((.(.((((.(((..((((((..(((((((((((((((.....))))))..(.....).)))))))..))..).)).)))..))).))))).)))).))).. ( -31.40) >DroYak_CAF1 71113 120 + 1 AUAAUUUAUUUUGAUCAGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGCGGAAACAUGUUUUGCUCAUCAAUGAAUUGGCUCUGAACCUGUUUGCCCUGCCUCAUUUA ............(((.((((.(.((((.(((..((((((..((((((((((((((......)))).((.....)).))))))))..))..).)).)))..))).))))).)))).))).. ( -28.40) >consensus AUAAUUUAUUUUGAUCAGGCCGCGGCAUGCAUUUUCGAUUGAUAUUGAUGGUUCCUUCUGGAGGAACAUGUUUUGCUCAUCAAUGAAUUGGCGCUGAACCUGUUUGCCCUGCCUCAUUUA ............(((.((((.(.((((.(((..((((.(..(((((((((((((((.....))))))..(.....).)))))))..))..)...))))..))).))))).)))).))).. (-27.92 = -28.36 + 0.44)



| Location | 12,704,803 – 12,704,923 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -24.76 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12704803 120 - 23771897 UAAAUGAGGCAGGGCAAACAGGUUCAGCGCCAAUUCAUUGAUGAGCAAAACAUGUUCCUCCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAU ...((.((((.(((((..(((((.....))).....(((((((..........((((((.....)))))))))))))............))..)))).).)))).))............. ( -27.30) >DroSec_CAF1 68445 120 - 1 UAAAUGAGGCAGGGCAAACAGGUUCAGCGCCAAUUCAUUGAUGAGCAAAACAUGUUCCUCCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCAGAUCAAAAUAAAUUAU .......(((.(((((..(((((.....))).....(((((((..........((((((.....)))))))))))))............))..)))).).)))................. ( -25.70) >DroSim_CAF1 67198 120 - 1 UAAAUGAGGCAGGGCAAACAGGUUCAGCGCCAAUUCAUUGAUGAGCAAAACAUGUUCCUCCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCAGAUCAAAAUAAAUUAU .......(((.(((((..(((((.....))).....(((((((..........((((((.....)))))))))))))............))..)))).).)))................. ( -25.70) >DroEre_CAF1 70532 120 - 1 UAAAUGAGGCAGGGCAAACAGGUUCAGAGCCAAUUCAUUGAUGAGCAAAACAUGUUCCUGCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAU ...((.((((.(((((..((..(((.(((....)))(((((((..........((((((.....))))))))))))).......)))..))..)))).).)))).))............. ( -30.10) >DroYak_CAF1 71113 120 - 1 UAAAUGAGGCAGGGCAAACAGGUUCAGAGCCAAUUCAUUGAUGAGCAAAACAUGUUUCCGCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAU ...((.((((.(((((..((..(((.(((....)))(((((((((((.....))))(((......)))..))))))).......)))..))..)))).).)))).))............. ( -28.00) >consensus UAAAUGAGGCAGGGCAAACAGGUUCAGCGCCAAUUCAUUGAUGAGCAAAACAUGUUCCUCCAGAAGGAACCAUCAAUAUCAAUCGAAAAUGCAUGCCGCGGCCUGAUCAAAAUAAAUUAU .......(((.(((((..(((((.....))).....(((((((..........((((((.....)))))))))))))............))..)))).).)))................. (-24.76 = -24.80 + 0.04)

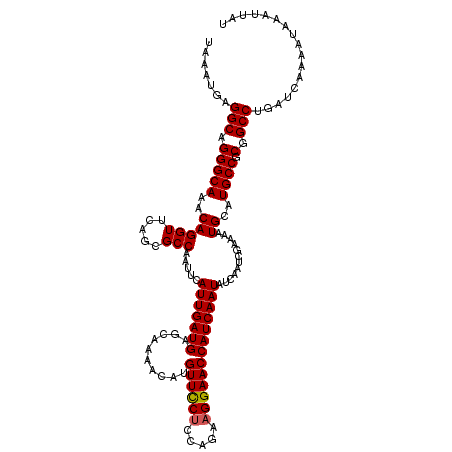
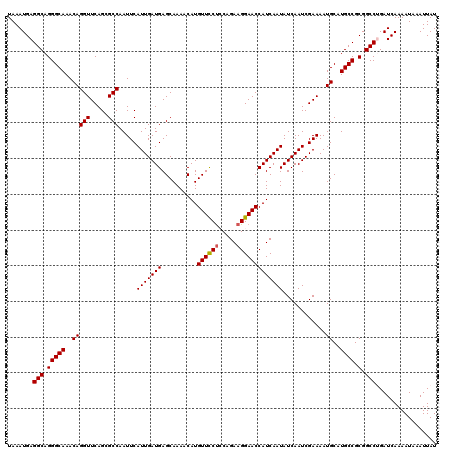
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:35 2006