| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,703,618 – 12,703,711 |
| Length | 93 |
| Max. P | 0.810827 |

| Location | 12,703,618 – 12,703,711 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.73 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -20.59 |
| Energy contribution | -20.46 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12703618 93 - 23771897 ---------------UGGCAGGACAU--AUCCACGGAGCGACACGUGUUGCGGAUGAGGACCAUCCGCCGAAAUGCACAGCCACAUCGCCUAAAUGCUGUGUAACUU-UUU ---------------.(((.(((...--.))).....(((((....)))))(((((.....))))))))....((((((((..............))))))))....-... ( -30.64) >DroSec_CAF1 67303 94 - 1 ---------------UGGCUGGCCGA--AUCCACAAAGCGACACGUGUUGCGGAUGAGGACCAUCCGCCGAAAUGCACAGCCACAUCGCCUAAAUGCUGUGUAACUUUUUU ---------------.((((((((..--((((.(((.(((...))).))).))))..)).)))...)))....((((((((..............))))))))........ ( -30.64) >DroSim_CAF1 66055 94 - 1 ---------------UGGCAGGCCGA--AUCCACGGAGCGACACGUGUUGCGGAUGAGGACCAUCCGCCGAAAUGCACAGCCACAUCGCCUAAAUGCUGUGUAACUUUUUU ---------------.(((...(((.--.....))).(((((....)))))(((((.....))))))))....((((((((..............))))))))........ ( -31.24) >DroEre_CAF1 69378 92 - 1 ---------------UGCGAGGACGC--CAGCGUCGAGCGACACGUGUUGCGGAUGAGGACCGUCCGUCGAAAUGCACAGCCAACUCGGCUAAAUGCUGUGUAACUU--UU ---------------..((((((((.--(..((((..(((((....))))).))))..)..))))).)))...((((((((..............))))))))....--.. ( -32.04) >DroYak_CAF1 69793 80 - 1 ---------------UGGAAGAA---------------GGACACGUGCUGCGGAUGAGGACCGUCCGUCGAAAUGCACAGCCACAUCGCCUAAAUGCUGUGUAACUU-UUU ---------------...(((((---------------(........(.(((((((.....))))))).)...((((((((..............)))))))).)))-))) ( -22.34) >DroPer_CAF1 62253 108 - 1 CUGGCCCUUCUUAACUUGAACGUCAACCGUCGACCAAGUGACACGUGUUGCGGAUGA--AACAUCCACGAAAAGGCACAGCAGCGUCCUGCAACCGCUUUGUAGUCU-CCC ..(((........(((((..((.(....).))..))))).(((((.((((((((((.--..)))))......(((.((......))))))))))))...))).))).-... ( -24.00) >consensus _______________UGGCAGGACGA__AUCCACCGAGCGACACGUGUUGCGGAUGAGGACCAUCCGCCGAAAUGCACAGCCACAUCGCCUAAAUGCUGUGUAACUU_UUU ....................((...............(((((....)))))(((((.....))))).))....((((((((..............))))))))........ (-20.59 = -20.46 + -0.14)

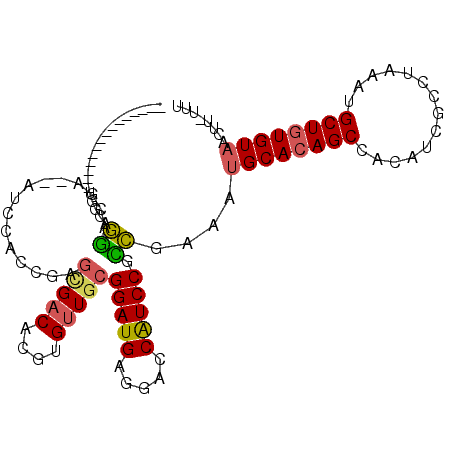

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:27 2006