| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,261,179 – 1,261,271 |
| Length | 92 |
| Max. P | 0.572408 |

| Location | 1,261,179 – 1,261,271 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
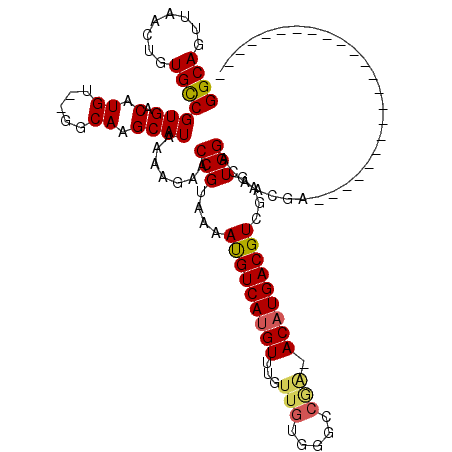
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -17.85 |
| Energy contribution | -17.93 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
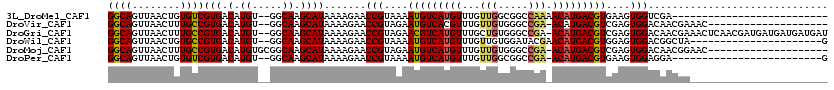
>3L_DroMel_CAF1 1261179 92 - 23771897 GGCAGUUAACUGUGUCGUGACAUGU--GGCAAGCAUAAAAGAACCGUAAAAUGUCAUGUUUGUUGGCGGCCAAAACAUGACGUGAAGUGGUCGA-------------------------- (((.((((((.....((((((((..--.((...............))...))))))))...)))))).)))......((((........)))).-------------------------- ( -24.36) >DroVir_CAF1 17210 97 - 1 GGCAGUUAACUUUGCCGUGACAUGU--GGCAAGCAUAAAAGAACCGUAGAAUGUCACGUUUGUUGUGGGCCGA-ACAUGACGUCGAGUGGACAACGAAAC-------------------- (((((......)))))(((((((..--.((...............))...))))))).((((((((..(((((-........))).))..))))))))..-------------------- ( -28.26) >DroGri_CAF1 27747 117 - 1 GGCAGUUAACUUUGCCGUGACAUGU--GGCAAGCAUAAAAGAACCGUAGAACGUCAUGUUUGCUGUGGGCCGA-ACAUGACGUCGAGUGGACAACGAAACUCAACGAUGAUGAUGAUGAU (((((......))))).....((((--.....))))........(((.((.((((((((((((.....).)))-))))))))))((((..........)))).))).............. ( -30.30) >DroWil_CAF1 24286 96 - 1 GGCAGUUAACUGUGCCGUGACAUGU--GGCAAGCAUAAAAGAACCGUAAAAUGUCAUGUUUGUUGUGGAUACGAACAUGACGUGGAGUGGACGGCUA----------------------G .((((....))))(((((...((((--.....)))).......(((....(((((((((((((.......)))))))))))))....))))))))..----------------------. ( -32.80) >DroMoj_CAF1 18137 99 - 1 GGCAGUUAACUUUGCCGUGACAUGUGCGGCAAGCAUAAAAGAACCGUAGAAUGUCAUGUUUGUUGUGGGCCGA-ACAUGACGUCGAGUGGACAACGGAAC-------------------- (((((......)))))......(((((.....)))))......((((.....((((((((((........)))-)))))))(((.....))).))))...-------------------- ( -32.70) >DroPer_CAF1 22742 92 - 1 GGCAGUUAACUGUGUCGUGACAUGU--GGCAAGCAUAAAAGAACCGUAAAAUGUCAUGUUUGUUGGCGGCCGA-ACAUGACGUGAAGUGGAGGA-------------------------G .((((....))))((((.......)--))).............(((....((((((((((((........)))-)))))))))....)))....-------------------------. ( -22.80) >consensus GGCAGUUAACUGUGCCGUGACAUGU__GGCAAGCAUAAAAGAACCGUAAAAUGUCAUGUUUGUUGUGGGCCGA_ACAUGACGUCGAGUGGACAACGA_______________________ ((((........))))(((.(.((.....)).)))).......(((....(((((((((...(((.....))).)))))))))....))).............................. (-17.85 = -17.93 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:39 2006