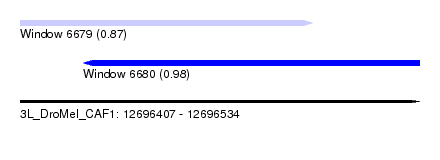
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,696,407 – 12,696,534 |
| Length | 127 |
| Max. P | 0.980036 |

| Location | 12,696,407 – 12,696,500 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -18.97 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12696407 93 + 23771897 ACAGGCUGAAGGCUCAUUAAGGCGCUCGACGGUGGCGUGGG----CGCGGGGCGU-UUACCGCGAUUCGGUUGCAAUUUCCAUAUGGAAAUUGAAUUU ...(((((((.((((((...(.((((....)))).))))))----)((((.....-...))))..))))))).((((((((....))))))))..... ( -34.40) >DroSec_CAF1 60127 95 + 1 ACAGGCUGAAGGCUCAUUAAGGCGUUCGAUGGUGGGGUGGGG---GGCGGGGCCUGUUACCGCGAUUCGGUUGCAAUUUCCAUAUGGAAAUUGAAUUU ...(((((((.(((......))).)))((...((.(((((.(---(((...)))).))))).))..)))))).((((((((....))))))))..... ( -33.00) >DroSim_CAF1 58857 95 + 1 ACAGGCUGAAGGCUCAUUAAGGCGUUCGAUGGUGGGGUGGGG---GGCGGGGCCUGUUACCGCGAUUCGGUUGCAAUUUCCAUAUGGAAAUUGAAUUU ...(((((((.(((......))).)))((...((.(((((.(---(((...)))).))))).))..)))))).((((((((....))))))))..... ( -33.00) >DroEre_CAF1 62435 80 + 1 ACAGGCUGAAGGCUCAUUAGGGCGAUCG------------------GCGGAGGCGGGAACCGCGAUUCGGUUGCAAUUUCCAUAUGGAAAUUGAAUUU ....(((((..((((....))))..)))------------------))(((.((((...))))..))).....((((((((....))))))))..... ( -27.80) >DroYak_CAF1 62573 98 + 1 ACAGGCUGAAGGCUCAUUAAGGCGAUCGGUGGCGGAGGAGGAGGCGGCGGAGGCGGGAACCGCGAUUCGGUUGCAAUUUCCAUAUGGAAAUUGAAUUU ...(((((((.(((......)))..((.((.((..........)).)).)).((((...))))..))))))).((((((((....))))))))..... ( -28.50) >DroPer_CAF1 55689 77 + 1 AU-GGCA-GAGUCUCAUUAAAGUGUU-GGCGCAC-------A---GGCGG--------AUCCAGGCUCGACUGCAAUUUCCAUAUGGAAAUUGAAUUU ..-((..-((((((.......((((.-...))))-------.---((...--------..))))))))..)).((((((((....))))))))..... ( -21.20) >consensus ACAGGCUGAAGGCUCAUUAAGGCGUUCGAUGGUGG_GUGGG____GGCGGGGCCUGUUACCGCGAUUCGGUUGCAAUUUCCAUAUGGAAAUUGAAUUU ...(((((((.(((......))).......................((((.........))))..))))))).((((((((....))))))))..... (-18.97 = -19.58 + 0.61)



| Location | 12,696,427 – 12,696,534 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -18.36 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12696427 107 - 23771897 GCAUCAAUUGG--GAAAACCACAUCAAGGCCGGUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACCGAAUCGCGGUAA-ACGCCCCGCG----CCCACGCCACCGUCGAGCGCC ((.((...(((--.....)))......(((.(((((.....((((((((....)))))))).........(((((...-.....)))))----......))))))))))))... ( -33.40) >DroSec_CAF1 60147 108 - 1 GCAUCA-UUGG--GAAAACCACAUCAAGGCCGAUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACCGAAUCGCGGUAACAGGCCCCGCC---CCCCACCCCACCAUCGAACGCC ((....-.(((--.....)))......(..((((((.....((((((((....))))))))..((((.....))))....(((...)))---..........))))))..))). ( -29.00) >DroSim_CAF1 58877 108 - 1 GCAUCA-UUGG--GAAAACCACAUCAAGGCCGAUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACCGAAUCGCGGUAACAGGCCCCGCC---CCCCACCCCACCAUCGAACGCC ((....-.(((--.....)))......(..((((((.....((((((((....))))))))..((((.....))))....(((...)))---..........))))))..))). ( -29.00) >DroEre_CAF1 62455 93 - 1 ACAUCA-UUGG--GAAAACCACAUCAAGGCCGGUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACCGAAUCGCGGUUCCCGCCUCCGC------------------CGAUCGCC ......-.(((--.....)))......((((((((((....((((((((....))))))))..........((((...)))).)))))------------------))...))) ( -30.00) >DroYak_CAF1 62593 111 - 1 ACAUCA-UUGG--GCAAACCACAUCAAGGCCGGUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACCGAAUCGCGGUUCCCGCCUCCGCCGCCUCCUCCUCCGCCACCGAUCGCC .....(-((((--.............((((.((((((....((((((((....))))))))..........((((...)))).))))))))))............))))).... ( -32.31) >DroPer_CAF1 55707 94 - 1 GGUGCU-GUGGCAGAAAACCGCAUCAAGGCCAGCGAAAAUUCAAUUUCCAUAUGGAAAUUGCAGUCGAGCCUGGAU--------CCGCC---U-------GUGCGCC-AACACU (((((.-..(((.(.....)(.(((.((((...(((.....((((((((....))))))))...))).)))).)))--------.))))---.-------..)))))-...... ( -33.80) >consensus GCAUCA_UUGG__GAAAACCACAUCAAGGCCGGUGGAAAUUCAAUUUCCAUAUGGAAAUUGCAACCGAAUCGCGGUAACAGGCCCCGCC____CCCAC_CCACCAUCGAACGCC ...........................(((((((((.....((((((((....))))))))..((((.....))))..........................))))))...))) (-18.36 = -18.70 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:24 2006