
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,688,491 – 12,688,609 |
| Length | 118 |
| Max. P | 0.864702 |

| Location | 12,688,491 – 12,688,587 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 87.03 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -23.18 |
| Energy contribution | -23.23 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
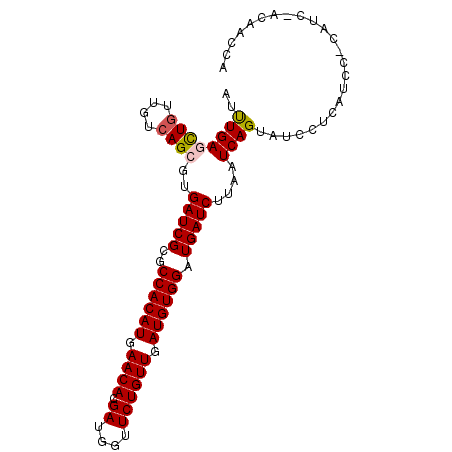
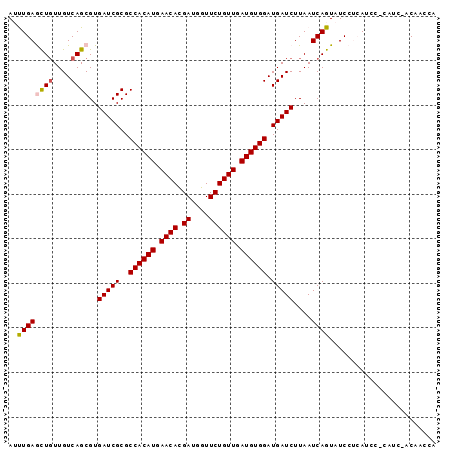
>3L_DroMel_CAF1 12688491 96 + 23771897 AUUUGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCCUCAUCCACUUCCACAUCCA ....((((((((((((((((....))))....)))...)))))))))(((.((.((((((((...............)))))))).)).))).... ( -26.86) >DroPse_CAF1 44994 96 + 1 AACUGAAUUGUUGUCAGCGUGAUCGCGCCACAUGAACAUGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCACAAAAGGCAUCGUCUACGA .(((((.(((..(((((((....))).((((((.((((.((....)))))).)))))).))))..))))))))........((((...)))).... ( -27.40) >DroSec_CAF1 52182 90 + 1 AUUUGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCCUCAUCC------ACAACCA ..((((((((....))))..(((((..((((((.((((.((....)))))).)))))).)))))....))))...........------....... ( -25.50) >DroSim_CAF1 50991 90 + 1 AUUUGAGCUCUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCCUCAUCC------ACAUCCA ....(((((.((((((((((....))))....)))...))).)))))....(((((((((((...............))))))------))))).. ( -25.76) >DroYak_CAF1 54451 96 + 1 AUUUGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCCUCAUCCACAUCCACUUCCA ....((((((((((((((((....))))....)))...))))))))).((.(((((((((((...............))))))))))).))..... ( -30.06) >DroPer_CAF1 48636 96 + 1 AACUGAAUUGUUGUCAGCGUGAUCGCGCCACAUGAACAUGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCACAAAAGGCAUCGUCUACGA .(((((.(((..(((((((....))).((((((.((((.((....)))))).)))))).))))..))))))))........((((...)))).... ( -27.40) >consensus AUUUGAGCUGUUGUCAGCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCCUCAUCC_CAUC_ACAACCA ..((((((((....))))..(((((..((((((.((((.((....)))))).)))))).)))))....))))........................ (-23.18 = -23.23 + 0.06)

| Location | 12,688,507 – 12,688,609 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -19.73 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
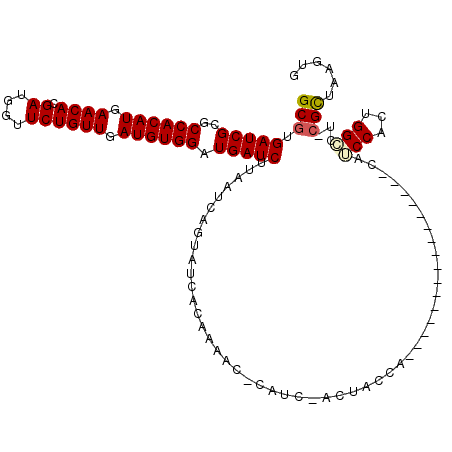
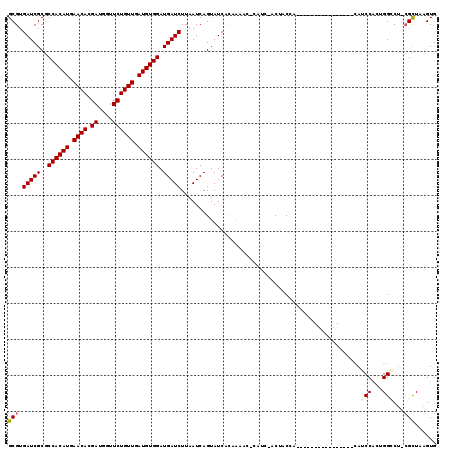
>3L_DroMel_CAF1 12688507 102 + 23771897 GCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCCUCAUCCACUUCCACAUCCA----------------CAUCCACUGGGCU-CGCUAAGUG ((((....))))...........((((....(((.((.((((((((...............)))))))).)).)))....----------------))))((((.((..-..)).)))) ( -26.86) >DroPse_CAF1 45010 117 + 1 GCGUGAUCGCGCCACAUGAACAUGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCACAAAAGGCAUCGUCUACGAAGGACGUAGAGCGAGCCAGCCA-GGGCCU-CGUUAAGUG ..(((((.((.((((((.((((.((....)))))).)))))).((((....)))))))))))....(((.(((((((((.....)))))).)))))).((..-..))..-......... ( -39.20) >DroSec_CAF1 52198 96 + 1 GCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCCUCAUCC------ACAACCA----------------CAUCCACUGGGCU-CGCUAAGUG (((....)))(((.((.((((......))))(((.(.(((((((((...............))))))------))).).)----------------)).....))))).-......... ( -24.86) >DroYak_CAF1 54467 102 + 1 GCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCCUCAUCCACAUCCACUUCCA----------------CAUCCACUGGCCU-CGCUAAGUG ((((....))))...........((((.....((.(((((((((((...............))))))))))).)).....----------------))))(((((((..-.))).)))) ( -30.06) >DroAna_CAF1 53433 97 + 1 GCGUGAUCGCGCCACAUGAACACGAUGAUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCACAGAAG------GCCUCUC----------------UAUCCUCCGGCCAGGGCUAAGUG (((((((.((.((((((.((((.((....)))))).)))))).((((....)))))))))))....(------(((....----------------........))))...))...... ( -29.80) >DroPer_CAF1 48652 117 + 1 GCGUGAUCGCGCCACAUGAACAUGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCACAAAAGGCAUCGUCUACGAAGGACGUAGAGCGAGCCAGCCA-GGGCCU-CGUUAAGUG ..(((((.((.((((((.((((.((....)))))).)))))).((((....)))))))))))....(((.(((((((((.....)))))).)))))).((..-..))..-......... ( -39.20) >consensus GCGUGAUCGCGCCACAUGAACACGAUGGUUCUGUUGAUGUGGAUGAUCUUAAUCAGUAUCACAAAAC_CAUC_ACUACCA________________CAUCCACUGGCCU_CGCUAAGUG (((.(((((..((((((.((((.((....)))))).)))))).)))))..................................................(((...)))...)))...... (-19.73 = -19.78 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:22 2006