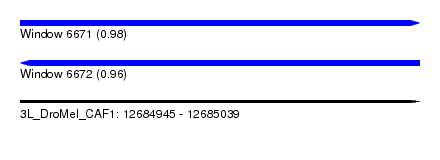
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,684,945 – 12,685,039 |
| Length | 94 |
| Max. P | 0.977465 |

| Location | 12,684,945 – 12,685,039 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.45 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -12.23 |
| Energy contribution | -12.06 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12684945 94 + 23771897 AACUUAUGAAUGGAGUUA---ACAAAAGGCGGGGUAUAU--------UGCCAGAG--------UGACUGGAGUGAGCCGGGGGUUAAUAAACCCUACACAAGUUUUUUAUGGU (((((.......))))).---..(((((((..(((.(((--------(.((((..--------...)))))))).))).((((((....))))))......)))))))..... ( -23.20) >DroPse_CAF1 41490 83 + 1 AAAUUAUGAAUGGCACUAGUGGCAAAAGGUGGU--------------------AA--------CUUCUCUGGGGAGCCAGAGGUUAAUAAACCUUU--UAAGUUAUUAAUGCU ...........((((.((((((((((((((..(--------------------((--------(..((((((....))))))))))....))))))--)..))))))).)))) ( -28.60) >DroSec_CAF1 48735 99 + 1 AACUUAUGAAUGGAGUUA---ACAAAAGGUGGGUUAUAUG-GUAUAUGGCCAGAG--------GGACUGGAGUGAGCCGGGGGUUAAUAAACCCUA--CGAGUUAUUUAUGGU ...((((((((((.((..---.................((-((.(((..((((..--------...)))).))).))))((((((....)))))))--)...)))))))))). ( -22.40) >DroSim_CAF1 47524 86 + 1 AACUUAUGAAUGGAGUUA---ACAAAAGGCGGGUUAUAUG-GU---------------------GACUGGAGUGAGCCGGGGGUUAAUAAACCCUA--CAAGUUAUUUAUGGU ...((((((((((.....---........(((.((((..(-..---------------------..)....)))).)))((((((....)))))).--....)))))))))). ( -20.50) >DroEre_CAF1 49348 80 + 1 AACUUAUGAAUGGAGGUA---ACAAAAGG--------------------CCAGAA--------UGACUGGAGUGAGCCGGGGGUUAAUAAACCCCA--CAGGUUAUUUAUGGU ...((((((((.......---.((.....--------------------((((..--------...))))..))((((.((((((....)))))).--..)))))))))))). ( -21.80) >DroYak_CAF1 50508 108 + 1 AACUUAUGAAUGGAGUUA---ACAAAAGGCGGUAUACACAUACAUAUGGCCAGAGUGGAAACUUGACUGUAGUGAGCCGGGGGUUAAUAAACCCCA--CCAGUUAUUUAUGGU ..(((((...((.((((.---......(((.((((........)))).))).((((....)))))))).)))))))...((((((....))))))(--(((........)))) ( -29.30) >consensus AACUUAUGAAUGGAGUUA___ACAAAAGGCGGG_UAUAU__________CCAGAG________UGACUGGAGUGAGCCGGGGGUUAAUAAACCCUA__CAAGUUAUUUAUGGU ....(((((((((..............(((.............................................))).((((((....)))))).......))))))))).. (-12.23 = -12.06 + -0.17)



| Location | 12,684,945 – 12,685,039 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 71.45 |
| Mean single sequence MFE | -17.42 |
| Consensus MFE | -6.86 |
| Energy contribution | -6.92 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12684945 94 - 23771897 ACCAUAAAAAACUUGUGUAGGGUUUAUUAACCCCCGGCUCACUCCAGUCA--------CUCUGGCA--------AUAUACCCCGCCUUUUGU---UAACUCCAUUCAUAAGUU .........((((((((..(((((....)))))..(((.....((((...--------..))))..--------.........)))......---..........)))))))) ( -17.99) >DroPse_CAF1 41490 83 - 1 AGCAUUAAUAACUUA--AAAGGUUUAUUAACCUCUGGCUCCCCAGAGAAG--------UU--------------------ACCACCUUUUGCCACUAGUGCCAUUCAUAAUUU .((((((......((--((((((....((((((((((....))))))..)--------))--------------------)..))))))))....))))))............ ( -20.50) >DroSec_CAF1 48735 99 - 1 ACCAUAAAUAACUCG--UAGGGUUUAUUAACCCCCGGCUCACUCCAGUCC--------CUCUGGCCAUAUAC-CAUAUAACCCACCUUUUGU---UAACUCCAUUCAUAAGUU ........((((...--..(((((....)))))..((......((((...--------..)))).......)-)................))---))................ ( -12.92) >DroSim_CAF1 47524 86 - 1 ACCAUAAAUAACUUG--UAGGGUUUAUUAACCCCCGGCUCACUCCAGUC---------------------AC-CAUAUAACCCGCCUUUUGU---UAACUCCAUUCAUAAGUU .........((((((--(.(((((....)))))..((((......))))---------------------..-...................---...........))))))) ( -12.80) >DroEre_CAF1 49348 80 - 1 ACCAUAAAUAACCUG--UGGGGUUUAUUAACCCCCGGCUCACUCCAGUCA--------UUCUGG--------------------CCUUUUGU---UACCUCCAUUCAUAAGUU ........(((((((--.((((((....)))))))))......((((...--------..))))--------------------......))---))................ ( -16.70) >DroYak_CAF1 50508 108 - 1 ACCAUAAAUAACUGG--UGGGGUUUAUUAACCCCCGGCUCACUACAGUCAAGUUUCCACUCUGGCCAUAUGUAUGUGUAUACCGCCUUUUGU---UAACUCCAUUCAUAAGUU ......(((((..((--(((((((....)))))).((..((((((((((((((....))).))))....)))).)))....)))))..))))---)................. ( -23.60) >consensus ACCAUAAAUAACUUG__UAGGGUUUAUUAACCCCCGGCUCACUCCAGUCA________CUCUGG__________AUAUA_CCCGCCUUUUGU___UAACUCCAUUCAUAAGUU ...................(((((....)))))..((((......))))................................................................ ( -6.86 = -6.92 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:17 2006