| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 12,679,112 – 12,679,260 |
| Length | 148 |
| Max. P | 0.697482 |

| Location | 12,679,112 – 12,679,220 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.32 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -16.79 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12679112 108 + 23771897 AACCAAACAACUAAUCAGAGUGUUAUCUUUUGGGGUU-UUU-------UUUCUUUUCAACAGGCUGGCAUCGAUCUGAAGGAGAACCUGGUGGCCGGAGCAUCGCCCUGGCCAUAU .(((..........((((((((((((((.(((..(..-...-------.....)..))).))).))))))...)))))(((....))))))((((((.((...)).)))))).... ( -30.60) >DroVir_CAF1 51942 93 + 1 AUGCCAAA-GUUAAU-AG---AUUAUGCUU-----UU-AU------UCGAUUG------CAGGCCGGCAUUGAUUUAAAGGAGAACCUGGUGGCCGGAGCUGCACCAUGGCCGUAU ..((((.(-((((((-((---(.......)-----))-))------).))))(------(((.(((((..........(((....)))....)))))..))))....))))..... ( -24.44) >DroSec_CAF1 42961 115 + 1 AACCAAACAACUAAUCAGAGCGUUAUCUUUCGGGUUU-UUUUCUUUCUUUUUUUUUCAACAGGCUGGCAUCGAUCUGAAGGAGAACCUUGUGGCCGGAGCAUCGCCCUGGCCAUAU .................(((.(((.(((((((((((.-.(.((..(((.((......)).)))..)).)..))))))))))).))))))((((((((.((...)).)))))))).. ( -30.40) >DroSim_CAF1 41768 116 + 1 AACCAAACAACUAAUCAGAGCGUUAUCUUUCGGGUUUUUUUUCUUUCUUUUUUUUUCAACAGGCUGGCAUCGAUCUGAAGGAGAACCUGGUGGCCGGAGCAUCGCCCUGGCCAUAU ................(((......)))..(((((((((((((..((........................))...)))))))))))))((((((((.((...)).)))))))).. ( -28.76) >DroEre_CAF1 43584 87 + 1 AAUCAAAAUUCUAAU-----------CUUUGGGUUUU-UUUU-----------------CAGGCUGGCAUCGAUCUGAAGGAGAACCUGGUGGCCGGAGCAUCGCCCUGGCCAUAU ...............-----------....(((((((-((((-----------------(((..((....))..)))))))))))))).((((((((.((...)).)))))))).. ( -33.50) >DroYak_CAF1 44617 104 + 1 AAUCAAAAAUCUAAU-----------CUUUUGGGUUU-UUUUGUUUGUUUUCGUUCCAACAGGCUGGCAUCGAUCUGAAGGAGAACCUGGUGGCCGGAGCAUCGCCCUGGCCAUAU ....((((((((((.-----------...))))))))-)).............((((..(((..((....))..)))..))))......((((((((.((...)).)))))))).. ( -29.20) >consensus AACCAAAAAACUAAU_AG___GUUAUCUUUCGGGUUU_UUUU_____UUUUUUUUUCAACAGGCUGGCAUCGAUCUGAAGGAGAACCUGGUGGCCGGAGCAUCGCCCUGGCCAUAU ...........................................................(((..((....))..))).(((....))).((((((((.((...)).)))))))).. (-16.79 = -16.73 + -0.05)
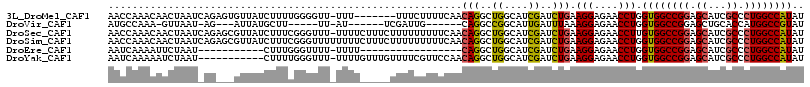
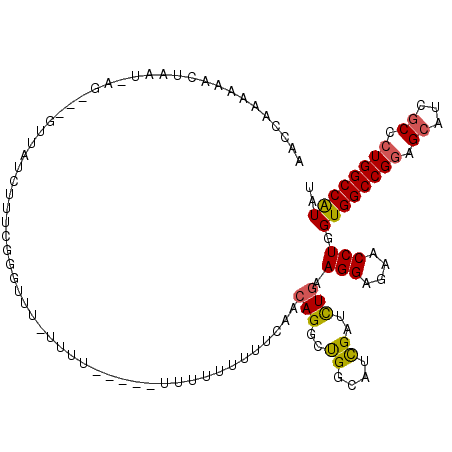

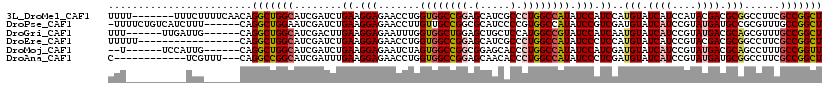
| Location | 12,679,148 – 12,679,260 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -26.14 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 12679148 112 + 23771897 UUUU-------UUUCUUUUCAACAGGCUGGCAUCGAUCUGAAGGAGAACCUGGUGGCCGGAGCAUCGCCCUGGCCAUAUCCAUCCAUGUAUCAUCCAUACGACGCGGCCUUCGCCGGCU ....-------.............(((((((...........(((.......((((((((.((...)).)))))))).....))).((..((........))..))......))))))) ( -33.60) >DroPse_CAF1 35270 112 + 1 -UUUUCUGUCAUCUUU------CAGGCUGGAAUCGAUCUGAAGGAGAACCUUGUUGCCGGCGCAUCCCCGUGGCCAUAUCCGUCGAUGUAUCAUCCGUAUGAUGCCGCGUUUGCCGGCU -......((..(((((------(((..((....))..))))))))..))......(((((((......((((((.(((((....)))))(((((....)))))))))))..))))))). ( -38.10) >DroGri_CAF1 25390 107 + 1 UUU------UUGAUUG------CAGGCUGGCAUCGACUUGAAGGAGAAUUUGGUGGCUGGAGCUGCUCCAUGGCCGUAUCCAUCAAUGUAUCAUCCGUAUGACGCAGCGUUUGCCGGCU ...------.......------..((((((((.((..((((.(((.....((((.(.(((((...)))))).))))..))).))))((..((((....))))..)).))..)))))))) ( -35.00) >DroEre_CAF1 43609 102 + 1 UUUUU-----------------CAGGCUGGCAUCGAUCUGAAGGAGAACCUGGUGGCCGGAGCAUCGCCCUGGCCAUAUCCCUCCAUGUAUCAUCCGUACGACGCGGCCUUCGCCGGCU .....-----------------..(((((((...........((((......((((((((.((...)).))))))))....)))).........((((.....)))).....))))))) ( -37.30) >DroMoj_CAF1 52478 105 + 1 --U------UCCAUUG------CAGGCUGGCAUCGAUCUGAAGGAGAAUCUAGUGGCCGGCGGAGCACCCUGGCCAUAUCCAUCGAUGUAUCAUCCGUAUGACGCAGCCUUUGCCGGUU --.------.((...(------(((((((((((((((.....(((.......((((((((.((....)))))))))).))))))))))).((((....))))..)))))..))).)).. ( -37.10) >DroAna_CAF1 43686 104 + 1 C------------UCGUUU---CAGGCCGGCAUCGAUUUGAAGGAGAACCUGGUGGCCGGAGCAACACCCUGGCCAUAUCCCUCGAUGUAUCAUCCGUAUGAUGCGGCCUUCGCCGGCU .------------......---..((((((((((((.....(((....))).((((((((.(.....).)))))))).....)))))(((((((....))))))).......))))))) ( -40.00) >consensus UUU__________UUG______CAGGCUGGCAUCGAUCUGAAGGAGAACCUGGUGGCCGGAGCAUCACCCUGGCCAUAUCCAUCGAUGUAUCAUCCGUAUGACGCGGCCUUCGCCGGCU ........................(((((((........((.(((.......((((((((.(.....).)))))))).))).))..(((.((((....)))).)))......))))))) (-26.14 = -26.58 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:14 2006